Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
African Journal of Laboratory Medicine
On-line version ISSN 2225-2010
Print version ISSN 2225-2002
Afr. J. Lab. Med. vol.12 n.1 Addis Ababa 2023
http://dx.doi.org/10.4102/ajlm.v12i1.2167
BRIEF REPORT
Antibacterial activity of soil-isolated Bacillus altitudinis/pumilus complex against methicillin-resistant Staphylococcus aureus from Mwanza, Tanzania
Reuben N. AbednegoI; Vitus SilagoII
IMicrobiology Laboratory, National Public Health Laboratory, Dar es Salaam, United Republic of Tanzania
IIDepartment of Microbiology and Immunology, Weill Bugando School of Medicine, Catholic University of Health and Allied Sciences, Mwanza, United Republic of Tanzania
ABSTRACT
Antimicrobial resistance in methicillin-resistant Staphylococcus aureus and beta-lactamase-producing Gram-negative bacteria is a global health concern necessitating research and the development of effective antimicrobial agents. This study, conducted in May 2020 in Mwanza, Tanzania, aimed to determine the antibacterial activity of metabolites from soil-isolated Bacillus species against clinical bacterial pathogens. One soil-isolated Bacillus species, identified as Bacillus altitudinis/pumilus complex, showed antibacterial activity against Gram-positive cocci, including a methicillin-resistant S. aureus strain with inducible clindamycin resistance, previously isolated from a patient with osteomyelitis. Bacillus altitudinis/pumilus complex metabolites may be a potential source of antimicrobial agents against multidrug-resistant bacteria.
WHAT THIS STUDY ADDS: The study supports existing research on the discovery and development of new antimicrobial agents against multi-drug-resistant bacteria. We report the antimicrobial activity of metabolites extracted from soil-isolated Bacillus altitudinis/pumilus complex strains against Gram-positive bacteria, including a methicillin-resistant Staphylococcus aureus strain with inducible clindamycin resistance.
Keywords: Bacillus altitudinis/pumilus complex; Gram-positive cocci; methicillin-resistant Staphylococcus aureus; discovery of new antibiotic agent; soil bacteria.
Introduction
The upsurge of multidrug-resistant (MDR) infections has led to an increase in healthcare costs and mortalities. Other negative impacts of MDR infections include prolonged days of hospitalisation and lack of prophylactic protection.1,2 Extended-spectrum beta-lactamase-producing Gram-negative bacteria, methicillin-resistant Staphylococcus aureus (MRSA) and inducible clindamycin-resistant S. aureus are common MDR bacteria reported globally.3,4,5 According to estimates, by 2050, MDR infections could cost up to $100 trillion United States dollars annually and cause approximately 10 million deaths per year.6,7 Moreover, a predictive statistical modelling study published in 2022 estimated that 1.27 million deaths were directly attributable to antimicrobial resistance (AMR), and 4.95 million deaths were associated with AMR globally in 2019.8 These deaths were largely due to MDR strains of Escherichia coli, S. aureus, and Klebsiella pneumoniae.8 These projections have led to urgent calls by the World Health Organization to guide and promote research and development of new effective antimicrobials against MDR strains.9 Since 1987 to date, no new class of antibiotics has been discovered and successfully introduced for clinical use, particularly for the treatment of infections caused by MDR bacteria such as extended-spectrum beta-lactamase-producing Gram-negative bacteria, MRSA and inducible clindamycin-resistant S. aureus strains.10
Currently, scientists are struggling to discover potential sources of antibiotics with activity against MDR bacteria. Various potential sources have been explored, including reptile blood,11 soil microorganisms,12 marine microorganisms,13 and plants.14 Certain microorganisms produce antimicrobial compounds such as antibiotics, antifungals, and antivirals as a means of gaining a competitive advantage in their environment where resources such as food are limited.15 The majority of antibiotics in clinical use today were sourced from microorganisms.16 Antimicrobials such as penicillin G (sourced from Penicillium notatum), vancomycin (Amycolatopsis orientalis), gentamicin (Micromonospora purpurea), and lincosamides (Streptomyces lincolnensis) are some examples that have been derived from microorganisms.16
This study aimed to test the antibacterial activity of Bacillus species isolated from soil samples in Mwanza, Tanzania, against medically important pathogenic bacteria, including MDR strains such as extended-spectrum beta-lactamase-producing Gram-negative bacteria, MRSA, and inducible clindamycin-resistant S. aureus isolated from clinical samples.
Methods
Ethical considerations
This study was approved by the joint Catholic University of Health and Allied Sciences and Bugando Medical Centre Research Ethics and Review Committee with research clearance certificate number: CREC/298/2023. The current study used bacteria species previously isolated from clinical samples at Bugando Medical Centre and stored at -80 °C in the Microbiology Laboratory at Catholic University of Health and Allied Sciences in Mwanza, Tanzania. Participants' informed consent forms were not applicable because archived bacteria were used. Moreover, patient-related information such as socio-demographic data were not used in the current study, to ensure data confidentiality.
Study design, duration, sampling and setting
This was a laboratory-based study conducted in May 2020 in Mwanza, Tanzania. Surface soil samples were collected in sterile Falcon™ 50 mL conical tubes (Corning, Glendale, Arizona, United States) from 20 different locations of the same site at Bugando Hill in Mwanza, Tanzania. Soil samples were sent to the Microbiology Laboratory at the Catholic University of Health and Allied Sciences and processed within 1 h of collection.
Isolation of Bacillus species from soil samples
Soil samples were serially diluted in 0.85% sterile saline as described previously17 before being inoculated onto 5% sheep blood agar (BA; HiMedia, Mumbai, India) plates. Briefly, 1 g of soil sample was suspended in 5 mL of sterile saline, which was then vortexed for 15 s and allowed to settle for 5 min. One millilitre of the resulting supernatant was mixed with 4 mL of sterile saline, vortexed for 15 s, and then allowed to settle for 5 min. This step was repeated three times to obtain five-fold serial dilutions. From the fifth dilution, 1 mL of supernatant was inoculated on a BA plate using the pour plate technique. Plates were incubated aerobically at 37 °C for 20 h. Presumptive Bacillus species colonies (large-sized, dry, flat, greyish to whitish, and beta-haemolytic colonies) were selected and sub-cultured on fresh BA plates to obtain pure colonies. These plates were incubated aerobically at 37 °C for 20 h. Gram staining was used for the preliminary identification of Bacillus species before further analysis.
Extraction of bacterial metabolites and antibacterial activity testing
A loopful (10 µL) of each presumptive Bacillus isolate was suspended in 1 mL of sterile nuclease-free water in an Eppendorf tube (1.5 mL safe lock microcentrifuge tube; Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany). The tubes were incubated at 50 °C in a heating block for 30 min with intermittent vortex-mixing for 15 s every 10 min. We selected 50 °C for the extraction of bacterial metabolites because this temperature is insufficient for the destruction of metabolites. After incubation, the tubes were centrifuged at 12 000 revolutions per minute for 10 min, after which 0.8 mL of the supernatant from each tube was transferred into sterile Eppendorf tubes. The supernatants containing metabolites from soil-isolated presumptive Bacillus species were used immediately for antibacterial activity determination.
The well diffusion method, as described by Yilmaz and colleagues,18 was used to test the antibacterial activity of the extracted metabolites against one isolate each of extended-spectrum beta-lactamase-producing and non-producing E. coli and K. pneumoniae, MRSA and methicillin-susceptible S. aureus, inducible clindamycin-resistant S. aureus, coagulase-negative staphylococci, and Streptococcus pyogenes. These bacterial pathogens were isolated from clinical samples of previous studies conducted in the same setting19,20 and archived at -80 °C in 20% glycerol stocks. The recovery of these bacterial strains was performed by subculture on BA plates which were incubated at 37 °C for 24 h. Bacterial suspensions of these test strains were prepared using sterile saline and adjusted to 0.5 McFarland standard solution (Remel, Lenexa, Kansas, United States). The bacterial suspensions were then inoculated onto the surface of Mueller Hinton agar (MHA; HiMedia, Mumbai, India) plates. Within 15 min of inoculation, wells of 6 mm in diameter were created in the inoculated plates using a cork borer (Sigma Aldrich, Darmstadt, Germany), and 100 µL of the suspension containing the extracted metabolites was then pipetted into the bored wells. The inoculated MHA plates were incubated in an upright position in ambient air at 37 °C for 20 h. The diameters of zones of inhibitions were measured in millimetres from the edge of the inhibition zone to the margin of Bacillus species metabolites. The complete absence of inhibition was referred to as 'negative antibacterial activity' and the presence of a clear zone of inhibition was referred to as 'positive antibacterial activity'. This experiment was performed in duplicate to ensure the validity of our results. In cases of positive antibacterial activity, an average zone of inhibition was calculated and recorded as the final result.
Identification of Bacillus species with positive antibacterial activity
Only one soil-isolated presumptive Bacillus species with a clear zone of inhibition ('positive antibacterial activity') was further identified at the species level. We first conducted biochemical identification tests, including catalase production, coagulase production, oxidase production, urease production, DNase production, indole production, citrate utilisation, lactose fermentation, hydrolysis of bile aesculin, and motility tests. We also tested the capacity of the isolates to grow on BA supplemented with 7% NaCl, as well as at higher incubation temperatures of 50 °C and 70 °C. The biochemical identification tests were not conclusive. For further species identification, we also used the VITEK MS (BioMérieux, Baden, Germany) system, which is an automated microbial identification system that uses the Matrix Assisted Laser Desorption Ionization Time-of-Flight technology. The latter was performed at the Microbiology Laboratory, National Public Health Laboratory in Dar es Salaam, Tanzania.
Data analysis
Laboratory data were documented and analysed using Microsoft Excel (Microsoft Corporation, Redmond, Washington, United States). Categorical data were expressed as percentages.
Results
A total of 20 soil samples were collected and processed for the isolation of Bacillus species. Out of the 20 soil samples processed, 16 (80.0%) showed growth of presumptive Bacillus species. Of the 16 presumptive Bacillus species isolated, only one (6.3%) isolate - S9D - showed antibacterial activity against Gram-positive bacteria only, including the MRSA strain, which was also inducible clindamycin-resistant (Table 1 and Figure 1).
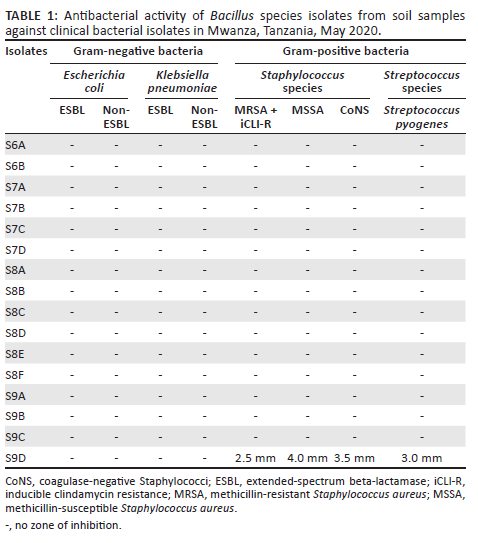
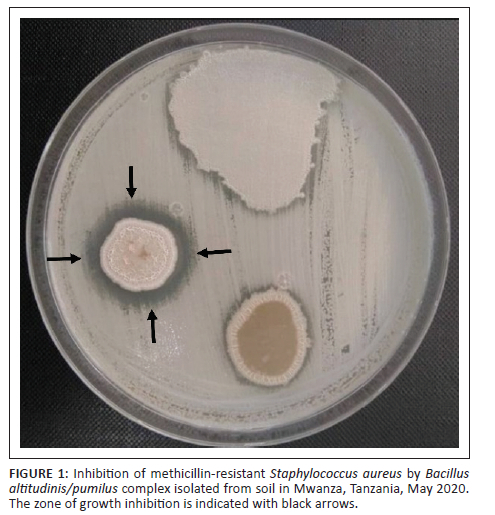
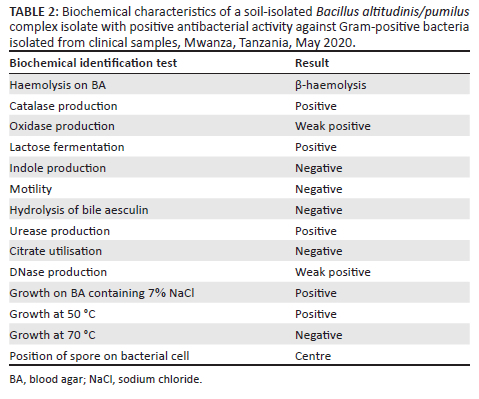
The presumptive Bacillus species isolate with positive antibacterial activity was Gram-positive and rod-shaped with central spores on microscopic examination after Gram staining. The isolate was also β-haemolytic on 5% sheep BA and demonstrated catalase production, sugar fermentation, growth on BA supplemented with 7% NaCl, and growth at 50 °C (Table 2). Using VITEK MS, the isolate was identified as Bacillus altitudinis/pumilus complex with a confidence level of 99.9%.
Discussion
In this study, we report a Bacillus altitudinis/pumilus complex isolate with antimicrobial activity against clinical Gram-positive bacterial strains, including an MRSA strain that was also inducible clindamycin-resistant. Due to the presence of several closely related species in the Bacillus genus, the VITEK MS system used in this study could not identify the Bacillus altitudinis/pumilus complex isolate at the species level.21 The study finding is similar to a previous study in Brazil in 2020,22 where marine-isolated Bacillus altitudinis/pumilus was shown to possess antimicrobial activity against MDR bacterial strains. Similarly, another study in Thailand in 2007 also reported that a soil-isolated Bacillus pumilus showed antibacterial activity against Gram-positive bacteria, including MRSA and vancomycin-resistant Enterococcus faecalis.23 The potential antimicrobial activity of Bacillus altitudinis/pumilus complex isolates has been linked to the production of bacilysins and bacteriocins.22 The report from Thailand in 2007 documented that pumilicin 4 is a bacteriocin produced by a Bacillus pumilus strain and had bactericidal activity against MRSA strains.23
We observed no antibacterial activity against Gram-negative bacteria, indicating that Bacillus altitudinis/pumilus complex may only be effective against Gram-positive bacteria. Similar findings were reported in Brazil in 2021 where a Bacillus altitudinis isolate from wetland sediment showed no antibacterial activity against Gram-negative bacteria.21 The presence of the outer membrane in Gram-negative bacteria limits cellular permeability,24 and may thus result in poor diffusion of antimicrobial metabolites into the cytoplasmic membrane for potential inhibition of bacterial growth.25
Limitations
Due to limited resources and funds, we were not able to identify the presumptive Bacillus species with negative antibacterial activity beyond the genus level. We also could not isolate, purify, or characterise the active metabolites from the Bacillus altitudinis/pumilus complex isolate with positive antibacterial activity.
Conclusion
Our findings suggest that metabolites from soil-isolated Bacillus altitudinis/pumilus complex can be a potential source of antimicrobial agents with activity against Gram-positive bacteria, including MRSA and inducible clindamycin-resistant strains.
Acknowledgements
Authors would like to express their sincerely appreciation to the Microbiology Laboratory at Catholic University of Health and Allied Sciences in Mwanza, Tanzania, and Microbiology Laboratory at National Public Health Laboratory in Dar es Salaam, Tanzania.
Competing interests
The authors declare that they have no financial or personal relationships that may have inappropriately influenced them in writing this article.
Authors' contributions
V.S. conceptualised the article. Both V.S. and R.N.A. performed sample collection, laboratory procedures, and prepared the article.
Sources of support
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability
All data presented in this article are available at Microbiology Laboratory at Catholic University of Health and Allied Sciences and can be accessed upon an official request to the corresponding author, V.S.
Disclaimer
The views and opinions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of any affiliated agency of the authors.
References
1.Friedman ND, Temkin E, Carmeli Y. The negative impact of antibiotic resistance. Clin Microbiol Infect. 2016;22(5):416-422. https://doi.org/10.1016/j.cmi.2015.12.002 [ Links ]
2.Wolf J. Antibiotic resistance threatens the efficacy of prophylaxis. Lancet Infect Dis. 2015;15(12):1368-1369. https://doi.org/10.1016/S1473-3099(15)00317-5 [ Links ]
3.Navidinia M. Detection of inducible clindamycin resistance (MLSBi) among methicillin-resistant Staphylococcus aureus (MRSA) isolated from health care providers. Arch Adv Biosci. 2015;6(1):91-96. [ Links ]
4.Yehouenou CL, Kpangon AA, Affolabi D, et al. Antimicrobial resistance in hospitalized surgical patients: A silently emerging public health concern in Benin. Ann Clin Microbiol Antimicrob. 2020;19:1-10. https://doi.org/10.1186/s12941-020-00398-4 [ Links ]
5.Giufrè M, Ricchizzi E, Accogli M, et al. Colonization by multidrug-resistant organisms in long-term care facilities in Italy: A point-prevalence study. Clin Microbiol Infect. 2017;23(12):961-967. https://doi.org/10.1016/j.cmi.2017.04.006 [ Links ]
6.O'Neill J. Review on antimicrobial resistance: Tackling a crisis for the health and wealth of nations. 2014. London: HM Government; 2016. [ Links ]
7.The World Bank. By 2050, drug-resistant infections could cause global economic damage on par with 2008 financial crisis. Washington, DC: World Bank; 2016. [ Links ]
8.Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399(10325):629-655. https://doi.org/10.1016/S0140-6736(21)02724-0 [ Links ]
9.World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. Geneva: WHO; 2017. [ Links ]
10.Silver LL. Challenges of antibacterial discovery. Clini Microbiol Rev. 2011;24(1):71-109. https://doi.org/10.1128/CMR.00030-10 [ Links ]
11.Bishop BM, Juba ML, Russo PS, et al. Discovery of novel antimicrobial peptides from Varanus komodoensis (Komodo dragon) by large-scale analyses and de-novo-assisted sequencing using electron-transfer dissociation mass spectrometry. J Proteome Res. 2017;16(4):1470-1482. https://doi.org/10.1021/acs.jproteome.6b00857 [ Links ]
12.Begum K, Mannan J, Rahman M, Mitchel-Antoine A, Opoku R. Identification of antibiotic producing bacteria from soil samples of Dhaka. J Microbiol Exp. 2017;4:00134. https://doi.org/10.15406/jmen.2017.04.00134 [ Links ]
13.Sosovele EM, Lyimo TJ, Hosea KM. In vitro antimicrobial activity of extracts from marine streptomyces isolated from mangrove sediments of Tanzania. J Biochem Technol. 2012;3(4):431-435. [ Links ]
14.Mwambete K. The in vitro antimicrobial activity of fruit and leaf crude extracts of Momordica charantia: A Tanzania medicinal plant. Afri Health Sci. 2009;9(1):34-39. [ Links ]
15.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8(1):15-25. https://doi.org/10.1038/nrmicro2259 [ Links ]
16.Rajeev L. Antibiotic discovery. Mat Meth. 2016;8:2671. https://doi.org/10.13070/mm.en.8.2671 [ Links ]
17.Chalasani AG, Dhanarajan G, Nema S, Sen R, Roy U. An antimicrobial metabolite from Bacillus sp.: Significant activity against pathogenic bacteria including multidrug-resistant clinical strains. Frontiers in Microbiol. 2015;6:1335. https://doi.org/10.3389/fmicb.2015.01335 [ Links ]
18.Yilmaz M, Soran H, Beyatli Y. Antimicrobial activities of some Bacillus spp. strains isolated from the soil. Microbiol Res. 2006;161(2):127-131. https://doi.org/10.1016/j.micres.2005.07.001 [ Links ]
19.Silago V, Kovacs D, Msanga DR, et al. Bacteremia in critical care units at Bugando Medical Centre, Mwanza, Tanzania: The role of colonization and contaminated cots and mothers' hands in cross-transmission of multidrug resistant Gram-negative bacteria. Antimicrob Resist Infect Control. 2020;9(1):1-14. https://doi.org/10.1186/s13756-020-00721-w [ Links ]
20.Silago V, Mushi MF, Remi BA, et al. Methicillin resistant Staphylococcus aureus causing osteomyelitis in a tertiary hospital, Mwanza, Tanzania. J Orthop Surg Res. 2020;15(1):1-6. https://doi.org/10.1186/s13018-020-01618-5 [ Links ]
21.Cavalini L, Jankoski P, Correa APF, Brandelli A, Motta AS. Characterization of the antimicrobial activity produced by Bacillus sp. isolated from wetland sediment. An Acade Bras Ciênc. 2021;93(Suppl 4):e20201820. https://doi.org/10.1590/0001-3765202120201820 [ Links ]
22.Freitas-Silva J, Silva-Oliveira T, Muricy G, Laport MS. Bacillus strains associated to homoscleromorpha sponges are highly active against multidrug resistant bacteria. Curr Microbiol. 2020;77(5):807-815. https://doi.org/10.1007/s00284-019-01870-x [ Links ]
23.Aunpad R, Na-Bangchang K. Pumilicin 4, a novel bacteriocin with anti-MRSA and anti-VRE activity produced by newly isolated bacteria Bacillus pumilus strain WAPB4. Curr Microbiol. 2007;55(4):308-313. https://doi.org/10.1007/s00284-006-0632-2 [ Links ]
24.Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794(5):808-816. https://doi.org/10.1016/j.bbapap.2008.11.005 [ Links ]
25.Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes. 2019;10(1):1-21. https://doi.org/10.1080/19490976.2018.1455790 [ Links ]
 Correspondence:
Correspondence:
Vitus Silago
vsilago.silago2@gmail.com
Received: 25 Jan. 2023
Accepted: 15 May 2023
Published: 18 July 2023














