Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
African Journal of Laboratory Medicine
On-line version ISSN 2225-2010
Print version ISSN 2225-2002
Afr. J. Lab. Med. vol.11 n.1 Addis Ababa 2022
http://dx.doi.org/10.4102/ajlm.v11i1.1482
ORIGINAL RESEARCH
Diagnostic challenges with accurate identification of Listeria monocytogenes isolates from food and environmental samples in South Africa
Teena S.M. ThomasI, II; Juno ThomasIII; Karren le RouxI, II; Sanelisiwe T. DuzeII; Faith MkhwanaziI; Adriano DuseI, II
IInfection Control Services Laboratory, National Health Laboratory Services, Johannesburg, South Africa
IIDepartment of Clinical Microbiology and Infectious Disease, School of Pathology, University of the Witwatersrand, Johannesburg, South Africa
IIICentre for Enteric Diseases, National Institute of Communicable Diseases, Johannesburg, South Africa
ABSTRACT
BACKGROUND: The 2017-2018 listeriosis outbreak in South Africa warranted testing for Listeria monocytogenes in food products and processing environments. Diagnostic tests are needed to accurately differentiate L. monocytogenes from other Listeria species.
OBJECTIVE: The study assessed the performance of the commonly used tests in our setting to accurately identify L. monocytogenes.
METHODS: The study was conducted in a public health laboratory in South Africa. Cultured isolates from food and environmental samples were tested both prospectively and retrospectively between August 2018 and December 2018. Isolates were phenotypically identified using tests for detecting β-haemolysis, Christie-Atkins-Munch-Peterson, alanine arylamidase (AlaA), mannosidase, and xylose fermentation. Listeria monocytogenes isolates were identified using automated systems, Microscan Walkaway Plus 96, Vitek® MS, Vitek® 2 and Surefast Listeria monocytogenes PLUS PCR. All results were compared to whole-genome sequencing results.
RESULTS: β-haemolysis and Christie-Atkins-Munch-Peterson tests gave delayed positivity or were negative for L. monocytogenes and falsely positive for one strain of Listeria innocua. The AlaA enzyme and Colorex Listeria agar lacked specificity for L. monocytogenes identification. Based on a few phenotypic test results, an aberrant L. monocytogenes strain and Listeria seeligeri strain were reported. All automated platforms overcalled L. monocytogenes in place of other Listeria species.
CONCLUSION: No test was ideal in differentiating Listeria species. This is an issue in resource-limited settings where these tests are currently used. Newer technologies based on enzyme-linked immunosorbent assay and other molecular techniques specific to L. monocytogenes detection need to be investigated.
Keywords: Listeria monocytogenes; food; environmental samples; diagnostic challenges; Africa.
Introduction
An extensive outbreak of listeriosis occurred in South Africa in 2017-2018. To date, it is the largest laboratory-confirmed Listeria monocytogenes foodborne outbreak described globally.1 The L. monocytogenes strain responsible for the outbreak was characterised on whole-genome sequencing (WGS) analysis as multi-locus sequence type 6.2 Consequently, wide-scale testing for L. monocytogenes in various food products and processing environments commenced. For appropriate food safety and public health interventions, the utilised diagnostic tests should reliably differentiate L. monocytogenes, the outbreak pathogen, from other Listeria species. Listeria species and L. monocytogenes share the same growth requirements and often coexist in the same environment; therefore, L. monocytogenes should be accurately discriminated from other co-occurring Listeria species.3
Six Listeria species (Listeria marthii, Listeria ivanovii, Listeria seeligeri, Listeria innocua, Listeria grayi and Listeria welshimeri) are closely related to L. monocytogenes. This close-relatedness challenges species differentiation.4 Although uncommon, 'atypical' strains, which do not exhibit typical phenotypic characteristics, of L. monocytogenes and other Listeria species have also been described.5
Several test methodologies are utilised to discriminate between the Listeria species; however, each has its pitfalls. L. monocytogenes is positive for the Christie-Atkins-Munch-Peterson (CAMP) test on sheep blood agar within 24 h of incubation.4 However, weakly haemolytic (showing haemolysis beyond 24 h of incubation) or non-haemolytic strains are frequently detected. Weak or no haemolysis is due to deletion of the hyl gene or its regulatory protein prfA, which regulates the expression of virulence factors required for L. monocytogenes pathogenesis.4 Other Listeria species, such as L. ivanovii (particularly with the CAMP test utilising Rhodococcus equi), L. seeligeri, and some L. innocua strains also show haemolytic capabilities which can make the utility of this test pointless.5,6,7
Listeria agar by Ottaviani and Agosti (Agar Listeria Ottaviani & Agosti medium, BioRad, Berkeley, California, United States) is recommended in the International Organization for Standardization 11290-1, 2017 standard for the isolation and differentiation of L. monocytogenes from other Listeria species.8 All Listeria species are selected for growth on the medium and produce blue-green colonies due to substrate degradation by β-D-glucosidase activity. L. monocytogenes and L. ivanovii can be differentiated from the other species due to the production of an opaque halo around the colonies as a result of phosphatidylinositol-specific phospholipase (PI-PLC) activity.9 The timing of the appearance of the opaque halo is also indicative of the species type. The halo is produced after 24 h incubation by L. monocytogenes and after 48 h of incubation by L. ivanovii.3,10 Other strains, such as L. seeligeri, L. welshimeri, and a few strains of L. innocua, may also possess the plcA gene, which codes for phospholipase activity that is responsible for creating the opaque halo around its colonies.4,5 In addition, other bacterial species, like Bacillus species, Cellulosimicrobium funkei, enterococci, Kochuria kristinae, Marinilactibacillus psychrotolerans, Rothia terrae, and coagulase-negative staphylococci, may also grow as blue-green colonies on Agar Listeria Ottaviani & Agosti medium.11 Bacillus circulans, Bacillus licheniformis, Enterococcus faecalis, Enterococcus faecium/durans, and Staphylococcus sciuri can produce a halo as well, which can make differentiation of L. monocytogenes from L. ivanovii difficult.11
The Analytical Profile Index Listeria test (BioMerieux, Marcy d'Etoile, France) fails in 10% - 15% of identification cases. The main reason for this failure is due to the weak colour determinations. This is particularly applicable to the arylamidase test. The arylamidase enzyme, tested for in the popular Differentiation Innocua Monocytogenes (DIM) test, is supposed to be negative in L. monocytogenes and positive in other Listeria species.12 Often a weak positive DIM result was considered a negative result, increasing the false positive L. monocytogenes determinations.4 This might be due to the doubtfulness of the colour determinations by the reader of the test. Furthermore, false negative identification in atypical L. monocytogenes strains is also frequent.4
Matrix-Assisted Laser Desorption Time of Flight (MALDI-TOF) mass spectrometry is a quick and easy methodology gaining popularity in several microbiology laboratories. However, MALDI-TOF reportedly misidentifies L. innocua as L. monocytogenes American Type Culture Collection (ATCC) strain and the L. seeligeri ATCC strain as L. monocytogenes or L. innocua.4
Rychert et al. reported that Vitek® Mass Spectrometer (MS) version 2.0 system (BioMerieux, Marcy d'Etoile, France) correctly identified only 76% (34/45) of L. monocytogenes to the species level and 9% (4/45) to the genus level, while in 15% (7/45) identification could not be finalised because split identification and re-testing were not performed.13 The Vitek® 2 system has also been reported to misidentify L. monocytogenes as L. innocua based on a negative reaction for phospholipase C in 1.4% (4/288) of a collection of isolates tested.14 The instrument could not identify an L. monocytogenes strain and gave a species error in another study.15 In a previous evaluation of the Vitek system, when genus level identification of various Listeria species was sought, the instrument had a sensitivity of 97.5%.16
The Microscan Walkaway Si system (Siemens Healthcare Diagnostics, West Sacramento, California, United States) could not identify one L. monocytogenes ATCC strain BAA-751 during a comparative study with the Vitek® 2 compact system.17 The reason for this was potentially attributed to the limited number of Listeria species strains on the database.
During the investigation of the South African listeriosis outbreak in 2017-2018, four Listeria species (L. monocytogenes, L. innocua, L. welshimeri, and L. seeligeri) were detected from food samples and environmental swabs tested at the Infection Control Services Public Health Laboratory in Johannesburg, South Africa. This is similar to what has been described elsewhere in outbreak settings.5 As a result, accurate discrimination of L. monocytogenes from other species is of critical importance. Whole-genome sequencing is a useful tool for confirmatory identification of L. monocytogenes and can be used as the reference standard test for comparing other tests.18
Subsequent to the reported limitations of Listeria tests commonly utilised in most public health laboratories, particularly in low- and middle-income countries, the Infection Control Services Public Health Laboratory evaluated the performance of the commonly utilised phenotypic tests (conventional phenotypic tests and chromogenic media) for the identification of Listeria species in comparison to WGS results. The Infection Control Services Public Health Laboratory also compared the performance of the different automated diagnostic systems available in the institution for the identification of L. monocytogenes utilising known Listeria isolates characterised by WGS.
The study results will inform whether current tests are acceptable for future use and, if not, it will justify the evaluation of other technologies for accurate identification of L. monocytogenes.
Methods
Ethical considerations
Only cultured isolates from food and environmental samples were utilised in this research. No isolates from animals or animal-derived samples were used. Therefore, no ethical clearance was required.
Study design and samples used
Data for this analysis were collected prospectively from August 2018 to December 2018 at the Infection Control Services Public Health Laboratory in Johannesburg. Isolates were cultured from food and environmental swabs of several food processing facilities across all of the provinces in South Africa during the listeriosis outbreak period. All Listeria isolates were identified on Vitek® 2 (BioMerieux, Marcy-I'Etoile, France). The phenotypic tests were performed either to (1) confirm the initial identification from Vitek® 2 or (2) to discriminate between Listeria species if two species identifications were given by Vitek® 2. As a result, not all phenotypic tests were performed on all isolates. The accuracy of the conventional phenotypic tests to discriminate the four Listeria species (L. monocytogenes, L. innocua, L. welshimeri, and L. seeligeri) was assessed (Table 1).3 The isolate identity (Vitek® 2 and phenotypic testing) was confirmed by WGS.

These isolates included 39 L. monocytogenes and 36 Listeria non-monocytogenes species, including 28 L. innocua, seven L. welshimeri, and one L. seeligeri.
Laboratory analyses
Beta (β)-haemolysis was performed on sheep blood agar. Plates were checked daily for up to 72 h. The CAMP test was performed using the Staphylococcus aureus ATCC strain 25923. The positive controls used for this test were Streptococcus agalactiae ATCC 13813 and L. monocytogenes ATCC 19115. The plates were examined daily for up to 72 h. The presence of arylamidase, mannosidase enzymes and acid production from D-xylose (dXYL) fermentation was assessed on the Vitek® 2 Gram-positive card based on the results of alanine arylamidase (AlaA), α-mannosidase (AMAN), and dXYL. The Vitek® 2 Gram-positive card was inoculated with the isolates as per the Vitek® 2 instrument training manual.19
The Colorex Listeria agar (E&O Laboratories Ltd, Bonnybridge, United Kingdom) has the same constituents as the Agar Listeria Ottaviani & Agosti medium. This medium was assessed and analysis was performed retrospectively using 59 of the 75 banked Listeria isolates from the analysis of the phenotypic tests. The isolates included 36 L. monocytogenes, 16 L. innocua, and seven L. welshimeri.
The laboratory also verified the performance of the automated platforms available. Analysis was performed retrospectively in December 2018 using 50 known Listeria isolates from the laboratory repository. The Listeria species included 20 L. monocytogenes strains and 30 Listeria species. The 30 Listeria species included 27 L. innocua, two L. seeligeri, and one L. welshimeri. These isolates were tested on four platforms, namely (1) Microscan Walkaway Plus 96 (Beckman Coulter Life Sciences, Indianapolis, Indiana, United States), (2) Vitek® MS version 3.0 system (BioMerieux, Marcy-I'Etoile, France), (3) Vitek® 2, and (4) Surefast Listeria monocytogenes PLUS polymerase chain reaction kit (Congen, Berlin, Germany), run on the Roche light cycler 2.0 (Roche, Basel, Switzerland) instrument. The Surefast kit identifies L. monocytogenes by amplifying a fragment of prfA and the detection limit of this assay is 10 CFU/mL as per our laboratory verification. Staff were blinded to the confirmatory WGS results of the isolates during the Colorex Listeria agar and automated systems assessments.
Data analysis
The sensitivity, specificity, positive predictive value, and negative predictive value of the test methods for detecting L. monocytogenes were calculated. Data were collected on Excel spreadsheets (Microsoft, Redmond, Washington, United States) and analysis was performed using two-by-two tables.20
Calculations were done as follows:
•Sensitivity = True positive L. monocytogenes isolates (Test and WGS positive) / Total WGS-confirmed L. monocytogenes isolates (True positive + False negative) × 100
•Specificity = True negative L. monocytogenes isolates (Test and WGS negative) / Total WGS-confirmed non-L. monocytogenes isolates (True negative + False positive) × 100
•Positive predictive value = True positive L. monocytogenes isolates (Test and WGS positive) / Total positive test results (True positive + False positive) × 100
•Negative predictive value = True negative L. monocytogenes isolates (Test and WGS negative) / Total negative test results (True negative + False negative) × 100
Results
The phenotypic results of the Listeria species in comparison to the WGS results are summarised in the Online Supplementary Table 1.
Performance of the phenotypic tests for L. monocytogenes identification
The three phenotypic tests used to confirm the identification of L. monocytogenes by Vitek® 2 and discriminate it from the other Listeria species were β-haemolysis, the CAMP test, and AlaA activity (Table 2).
Of the 39 L. monocytogenes isolates identified by WGS, all three phenotypic tests corroborated the WGS findings in 82% (32/39) of the isolates. β-haemolysis and the CAMP test were absent in 18% (7/39) of the isolates. Delayed positivity to both of these tests occurred at 72 h in 5.1% (2/39) of isolates. One L. innocua isolate was falsely positive to both of these tests.
Of the L. monocytogenes isolates, 18% (7/39) were falsely positive for AlaA on Vitek® 2. All L. innocua isolates and the one L. seeligeri isolate were falsely negative for AlaA.
Performance of the phenotypic tests for L. innocua identification
Of note, one out of the 28 L. innocua isolates was positive for both β-haemolysis and the CAMP test.
Performance of the phenotypic tests for L. welshimeri identification
The phenotypic tests used to identify L. welshimeri identified all seven of the isolates correctly. However, there was one isolate that had two identification options on Vitek® 2, namely L. monocytogenes and L. welshimeri. To discriminate between the two Listeria species, β-haemolysis, CAMP, and dXYL fermentation tests were assessed. The isolate was negative for β-haemolysis and the CAMP test but positive for dXYL fermentation, which suggested that the isolate is L. welshimeri. However, WGS results identified the isolate as L. monocytogenes. Performance indicators, such as sensitivity, specificity, positive predictive value, and negative predictive value, for the phenotypic tests (β-haemolysis, CAMP test and dXYL fermentation) differentiating L. welshimeri from L. monocytogenes were not done due to the low number of L. welshimeri isolates identified during the study period.
Performance of the phenotypic tests for L. seeligeri identification
One isolate was previously identified as L. monocytogenes based on Vitek® 2 identification (99% probability), positive β-haemolysis (at 24 h incubation), positive CAMP test (at 24 h incubation), negative AlaA enzyme activity, negative dXYL fermentation, and positive AMAN activity. However, WGS identified this isolate as L. seeligeri. Since there was only one L. seeligeri isolate, the performance of the tests (AlaA activity, AMAN activity, and dXYL fermentation) to discriminate this species from L. monocytogenes was not done.
Performance of the Colorex Listeria agar in the identification of L. monocytogenes
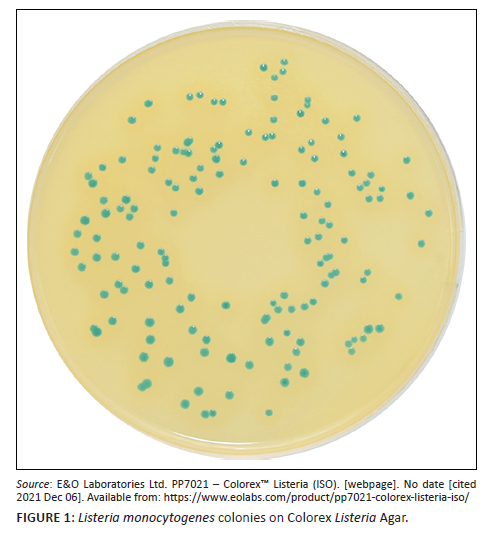
All 59 isolates representing the three Listeria species produced blue-green colonies on Colorex Listeria agar and 73% (43/59) of these isolates produced an opaque halo around the colonies (Figure 1).21 Of the 43 isolates that produced a halo, 84% (36/43) were identified as L. monocytogenes on WGS, while the remaining 16% (7/43) of isolates were identified as L. innocua (n = 5) and L. welshimeri (n = 2) on WGS (Online Supplementary Table 2). The sensitivity and specificity of the medium for accurate L. monocytogenes identification were 100% and 69%. The positive predictive value was 83.7% and negative predictive value was 100%.
Performance of the automated systems in the identification of L. monocytogenes
All the systems overcalled L. monocytogenes in place of other species (Table 3). All Listeria species results on the various automated systems are provided in Online Supplementary Table 3.
Discussion
From the evaluation, there was no ideal test for differentiating the Listeria species: all had limitations. β-haemolysis and the CAMP test are recommended to differentiate L. monocytogenes from L. innocua. However, these tests can give delayed positivity (up to three days later) or be negative for L. monocytogenes. In addition, they may be falsely positive for certain L. innocua strains. The DIM test for AlaA enzyme activity lacks specificity for L. monocytogenes detection. All of the other Listeria species also tested negative for this enzymatic activity, disproving its utility.
We report the first aberrant L. monocytogenes strain that fermented dXYL and an aberrant L. seeligeri strain that was negative for AlaA activity and dXYL fermentation and positive for AMAN activity. This further illustrates atypical strains that may potentially exist in our setting, complicating identification. Unfortunately, WGS could not be repeated on both of these isolates again to reconfirm the results and rule out the possibility of isolate mix-up.
The Colorex Listeria agar was able to correctly identify all L. monocytogenes isolates; however, it lacked specificity in discriminating other Listeria species from L. monocytogenes.
Limitations
There were also several shortcomings associated with the four automated diagnostic platforms tested. All platforms overcalled L. monocytogenes in place of the other Listeria species. This could be because these instruments were validated for clinical samples in which L. monocytogenes is the predominant species isolated. The Vitek® MS misidentified L. monocytogenes for other Listeria species and the Vitek® 2 gave both L. monocytogenes and L. innocua options for a few L. innocua isolates. The worst-performing platform for L. monocytogenes identification was Microscan and the best performer was the Surefast polymerase chain reaction kit.
Based on the above results, alternate testing platforms for L. monocytogenes identification need to be investigated.
Several other diagnostic methodologies are available to detect L. monocytogenes. These include (1) detection of L. monocytogenes by antibody-based assays, (2) molecular test methods such as Loop-mediated isothermal amplification (LAMP), DNA hybridisation or polymerase chain reaction utilising L. monocytogenes-specific gene targets that have been identified, and (3) the use of genetically engineered bacteriophages.3,7,9,12 Many of these assays are available as commercial automated kits approved by regulatory authorities. These technologies have been reported to perform well in the detection of L. monocytogenes. In addition, the automated systems are high throughput and significantly shortens the time to results of the traditional methods. The possible disadvantages of the above technologies would be: cost, staff expertise to perform the tests, and inhibition of tests (antibody-based and molecular) by the sample matrix. The molecular assays may also detect non-viable organisms.
Conclusion
We have demonstrated that the commonly used methodologies in most public health laboratories, particularly in low- and middle-income settings, are limited in differentiating L. monocytogenes from the other Listeria species. The accurate identification of L. monocytogenes is critical since it is the most predominant Listeria species causing human disease and, therefore, must not be missed by diagnostic tests. The large scale of this outbreak required upscaling laboratory support for public health sample testing. However, if the available systems in routine microbiology laboratories cannot discriminate between L. monocytogenes and the other Listeria species, overcalling or underreporting of L. monocytogenes can occur. Underreporting L. monocytogenes will prevent or delay identifying an outbreak source and promote its continuity with huge public health impact. Overcalling L. monocytogenes leads to the unnecessary closure of food production lines, which has huge financial implications for the company involved. Depending on the company's distribution level, halting production can also impact the community. In the outbreak setting, where L. monocytogenes prevalence in samples was comparatively higher, the positive predictive value of most of the tests assessed was unacceptable. Hence, in a non-outbreak setting, the performance of these tests will be worse.
Therefore, other technologies must be investigated for their discriminatory capabilities and accurate identification of L. monocytogenes in microbiology laboratories in low- and middle-income countries.
Acknowledgements
The authors would like to acknowledge the following for their role in the study: (1) the molecular and public health staff of the Infection Control Services Laboratory, National Health, Laboratory Services, for conducting the tests that were required, (2) the Microbiology Laboratory at Charlotte Maxeke Johannesburg Academic Hospital for granting permission to use the Vitek® 2 and Vitek® MS instruments, and (3) the National Institute of Communicable Diseases for providing whole-genome sequencing results for all the isolates used for this research.
Competing interests
The authors declare that they have no financial or personal relationships that may have inappropriately influenced them in writing this article.
Authors' contributions
T.S.M.T. and A.D. conceived the original idea for the study. T.S.M.T. supervised the study. F.M., S.T.D. and K.l.R. carried out the laboratory work and data collection. T.S.M.T. analysed the data. J.T. assisted with the whole-genome sequencing data and provided critical input to the article. All authors provided critical inputs to the manuscript.
Sources of support
This work was supported by the National Health Laboratory Services, South Africa, by providing all consumables used for this research.
Data availability
Derived data supporting the findings of this study are available from the corresponding author, T.S.M.T., on request.
Disclaimer
The views expressed in this publication are those of the authors and do not directly represent the views of any support agencies.
References
1.Smith AM, Tau NP, Shannon L, et al. Outbreak of Listeria monocytogenes in South Africa, 2017-2018: Laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog Dis. 2019;16(7):524-530. https://doi.org/10.1089/fpd.2018.2586 [ Links ]
2.Thomas J, Govender N, McCarthy KM, et al. Outbreak of listeriosis in South Africa associated with processed meat. NEJM. 2020;382(7):632-643. https://doi.org/10.1056/NEJMoa1907462 [ Links ]
3.Chen J-Q, Regan P, Laksanalamai P, Healey S, Hu Z. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci Hum Wellness. 2017;6(3):97-120. https://doi.org/10.1016/j.fshw.2017.06.002 [ Links ]
4.Pusztahelyi T, Szabó J, Dombrádi Z, Kovács S, Pócsi I. Foodborne Listeria monocytogenes: A real challenge in quality control. Scientifica. 2016;5768526:1-6. https://doi.org/10.1155/2016/5768526 [ Links ]
5.Orsi RH, Wiedmann M. Characteristics and distribution of Listeria species including Listeria species newly described since 2009. Appl Microbiol Biotechnol. 2016;100(12):5273-5287. https://doi.org/10.1007/s00253-016-7552-2 [ Links ]
6.Johnson J, Jinneman K, Stelma G, et al. Natural atypical Listeria innocua strains with Listeria monocytogenes pathogenicity Island 1 genes. Appl Environ Microbiol. 2004;70(7):4256-4266. https://doi.org/10.1128/aem.70.7.4256-4266.2004 [ Links ]
7.Gasanov U, Hughes D, Hansbro PM. Methods for the isolation and identification of Listeria species and Listeria monocytogenes: A review. FEMS Microbiol Rev. 2005;29(5):851-875. https://doi.org/10.1016/j.femsre.2004.12.002 [ Links ]
8.ISO 11290-1. Microbiology of the food chain- Horizontal method for the detection and enumeration of Listeria monocytogenes and of Listeria spp- Part 1: Detection method. 2nd ed. Geneva: Internation Standards Organization; 2017. [ Links ]
9.Law JW-F, Ab Mutalib N-S, Chan K-G, Lee L-H. An insight into the isolation, enumeration, and molecular detection of Listeria monocytogenes in food. Front Microbiol. 2015;6 (1227):1-15. https://doi.org/10.3389/fmicb.2015.01227 [ Links ]
10.Beumer RR, Hazeleger WC. Listeria monocytogenes: diagnostic problems. FEMS Immunol Med Microbiol. 2003;35(3):191-197. https://doi.org/10.1016/S0928-8244(02)00444-3 [ Links ]
11.Angelidis AS, Kalamaki MS, Georgiadou SS. Identification of non-Listeria species bacterial isolates yielding a β-D-glucosidase-positive phenotype on Agar Listeria according to Ottaviani and Agosti (ALOA). Int J Food Microbiol. 2015;193:114-129. https://doi.org/10.1016/j.ijfoodmicro.2014.10.022 [ Links ]
12.Janzten MM, Navas J, Corujo A, Moreno R, López V, Martínez-Suárez JV. Specific detection of Listeria monocytogenes in foods using commercial methods: From chromogenic media to real-time PCR. Span J Agric Res. 2006;4(3):235-247. https://doi.org/10.5424/sjar/2006043-198 [ Links ]
13.Rychert J, Burnham C-AD, Bythrow M, et al. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of gram-positive aerobic bacteria. J Clin Microbiol. 2013;51(7):2225-2231. https://doi.org/10.1128/jcm.00682-13 [ Links ]
14.De Lappe N, Lee C, O'Connor J, Cormican M. Misidentification of Listeria monocytogenes by the Vitek 2 system. J Clin Microbiol. 2014;52(9):3494-3495. https://doi.org/10.1128%2FJCM.01725-14 [ Links ]
15.Guo L, Ye L, Zhao Q, Ma Y, Yang J, Luo Y. Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification. J Thorac Dis. 2014;6(5):534-538. https://doi.org/10.3978%2Fj.issn.2072-1439.2014.02.18 [ Links ]
16.Odumeru JA, Steele M, Fruhner L, et al. Evaluation of accuracy and repeatability of identification of food-borne pathogens by automated bacterial identification systems. J Clin Microbiol. 1999;37(4):944-949. https://doi.org/10.1128/JCM.37.4.944-949.1999 [ Links ]
17.Rhoads S, Marinelli L, Imperatrice CA, Nachamkin I. Comparison of MicroScan WalkAway system and Vitek system for identification of gram-negative bacteria. J Clin Microbiol. 1995;33(11):3044-3046. https://doi.org/10.1128/jcm.33.11.3044-3046.1995 [ Links ]
18.Stasiewicz MJ, Oliver HF, Wiedmann M, et al. Whole genome sequencing allows for improved identification of persistent Listeria monocytogenes in food associated environments. Appl Environ Microbiol. 2015;81(17):6024-6037. https://doi.org/10.1128/aem.01049-15 [ Links ]
19.BioMerieux. Vitek 2 Systems: Vitek 2, XL, compact training manual. Biomerieux-South Africa local training manual, VT2C/ VT2/VT2 XL, V1-01/06/2014. Marcy-I'Etoile: BioMerieux; 2014. [ Links ]
20.Trevethan R. Sensitivity, specificity and predictive values: Foundations, pliabilities and pitfalls in research and practice. Front Public Health. 2017;307(5):1-7. https://doi.org/10.3389/fpubh.2017.00307 [ Links ]
21.E&O Laboratories Ltd webpage. PP7021 - Colorex™ Listeria (ISO). https://www.eolabs.com/product/pp7021-colorex-listeria-iso/. [ Links ]
 Correspondence:
Correspondence:
Teena Thomas
teena.thomas@nhls.ac.za
Received: 02 Dec. 2020
Accepted: 11 Feb. 2022
Published: 23 May 2022
Note: Additional supporting information may be found in the online version of this article as Supplementary Documents 1, 2 and 3.














