Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
African Journal of Laboratory Medicine
On-line version ISSN 2225-2010
Print version ISSN 2225-2002
Afr. J. Lab. Med. vol.7 n.2 Addis Ababa 2018
http://dx.doi.org/10.4102/ajlm.v7i2.806
BRIEF REPORT
Diagnosis of rifampicin-resistant tuberculosis: Discordant results by diagnostic methods
Winnie MwanzaI; Deborah MilimoI; Maureen M. ChilufyaI; Nkatya KaseseI; Maina C. LengweI; Stembiso MunkondyaI; Petra de HaasI, II; Helen AylesI, II; Monde MuyoyetaI
IZambia AIDS Related Tuberculosis (ZAMBART) Project, School of Medicine, University of Zambia, Lusaka, Zambia
IIDepartment of Infectious and Tropical diseases, London School of Hygiene and Tropical Medicine, Bloomsburg, London, United Kingdom
ABSTRACT
The performance of the Xpert© MTB/RIF and MTBDRplus assays for the detection of rifampicin resistant Mycobacterium tuberculosis was compared to culture-based drug susceptibility testing in 30 specimens with rifampicin-resistant and rifampicin-indeterminate Xpert MTB/RIF results collected between March 2012 and March 2014. Xpert MTB/RIF and MTBDRplus were 100% sensitive and 100% concordant for rifampicin resistance detection, but 3 of 13 samples (23%) positive for rifampicin resistance on Xpert MTB/RIF and MTBDRplus were negative for rifampicin resistance on mycobacteria growth indicator tube drug susceptibility testing. Specificity was 72% for Xpert MTB/RIF and 80% for MTBDRplus. Positive predictive value for Xpert MTB/RIF for multidrug resistant tuberculosis was 47.8% for new patients and 77.8% for previously treated patients; negative predictive value was 100% for both new and previously treated patients. The discordant rifampicin resistance test results indicate a need to fully characterise circulating rifampicin resistant Mycobacterium tuberculosis strains in Zambia and to inform the development of guidelines for decision-making in relation to diagnosis of drug-resistant tuberculosis.
Introduction
Management and control of multidrug resistant (MDR) tuberculosis, tuberculosis that is resistant to both isoniazid and rifampicin, relies on timely and correct diagnosis. The World Health Organization has endorsed the Xpert®MTB/RIF (Cepheid Sunnyvale, CA, United States) and GenoType MTBDRplus (HAIN Life Sciences, Nehren, Germany) tests for rapid diagnosis of MDR tuberculosis.1,2 For the Xpert MTB/RIF test, the current recommendation is to treat any rifampicin-resistant individual as a case of MDR tuberculosis until proven otherwise by subsequent drug susceptibility testing, after which treatment is to be tailored or individualised accordingly. The combination of Xpert MTB/RIF and MTBDRplus test results would be a powerful tool to achieve prompt confirmation of diagnosis of MDR tuberculosis. These genotypic methods, however, do not detect resistance conferred by mutations outside of the targeted gene region.3,4,5
We report comparative observations made in a pilot study on the performance of Xpert MTB/RIF, MTBDRplus and Mycobacteria Growth Indicator Tube (MGIT) drug susceptibility testing methods for detection of rifampicin resistance in Lusaka, Zambia, at the Zambart Central Laboratory (ZCL).
Methods
Ethical considerations
The study was approved by the University of Zambia biomedical research ethics committee (ref. no. 003-12-11) and all patients were requested to give written informed consent before participating in the study. The written informed consent was administered by study staff, and participants were given an opportunity to discuss the study before signing the informed consent form. To maintain confidentiality, each participant was allocated a unique study number that was used on all data collection tools, as well as for labelling of laboratory specimens. No names were used on any data collection tool. Informed consent forms and all study data were kept in secure cabinets with access limited to the principal investigator and the data manager. Paper-based case report forms were captured into secured, password-protected electronic databases.
Study Design
The study was conducted between March 2012 and March 2014 on samples collected during an evaluation of Xpert MTB/RIF use in primary health care facilities in Lusaka, Zambia.6,7 Samples were collected from consecutive patients presenting to the primary health care facility who had rifampicin-resistant and rifampicin-indeterminate Xpert MTB/RIF results for confirmation of rifampicin resistance and for quality assurance in the ZCL.
Samples were transported to the central laboratory on the same day of collection or the following day and no preservatives were added. At the ZCL, the samples were decontaminated using the NaOH/NaLC method (2% NaOH for 20 minutes) and 0.5 mL of the pellet was inoculated into each of two manual MGIT tubes for primary isolation and identification of M. tuberculosis.; 0.5 mL was used to repeat the Xpert MTB/RIF test, and two drops were used to prepare a concentrated smear for fluorescent microscopy. 1.5 mL of the Xpert MTB/RIF sample reagent was added to 0.5 mL of the re-suspended pellet and Xpert MTB/RIF testing was performed as per standard procedure described by the manufacture.
Acid-fast bacilli positive cultures were identified using the capillia tuberculosis Neo assay (TAUNS Laboratories, Inc., Shizuoka, Japan) to confirm presence of Mycobacterium tuberculosis complex.8 Confirmed M. tuberculosis complex isolates were tested for drug sensitivity using the Becton Dickinson manual MGIT SIRE (streptomycin, isoniazid, rifampicin, ethambutol; Becton Dickinson, Bergen County, New Jersey, United States) drug susceptibility testing method and the MTBDRplus assay (version 1; HAIN Life Sciences, Nehren, Germany). The drug concentrations used were 1.0 µg/mL streptomycin, 0.1 µg/mL isoniazid, 1.0 µg/mL rifampicin, and 5.0 µg/mL ethambutol. The MGIT SIRE tubes were read daily as per the manufacturer's recommendation. To perform the MTBDRplus assay, 1 mL of broth from an M. tuberculosis complex culture-positive MGIT tube was used for DNA extraction using the genolyse method. Briefly, 1 ml of culture broth was centrifuged, the pellet was incubated for 5 minutes at 95°C in lysis buffer after which a neutralisation buffer was added. Subsequent steps for polymerase chain reaction and hybridisation were followed as per standard procedure for MTBDRplus testing.9 All procedures included positive and negative controls.
The data were analysed using Stata version 13 (StataCorp LLC, College Station, Texas, United States). An initial descriptive analysis was done to determine the distribution of results, then the Xpert MTB/RIF results obtained at the primary health care setting were compared with the results obtained at the central laboratory. Further analysis was done to compare the performance of Xpert MTB/RIF, MTBDRplus and MGIT M. tuberculosis drug susceptibility testing and to determine the sensitivity and specificity of Xpert MTB/RIF and MTBDRplus compared to the gold standard MGIT culture for drug susceptibility testing.
Results
During the study period, 1070 patients had M. tuberculosis complex detected by the Xpert MTB/RIF test. Of these, 28 (2.6%) were positive for rifampicin resistance and 15 (1.4%) had indeterminate rifampicin resistance results. Of the 43 patients eligible for inclusion in the study (i.e. had rifampicin-resistant or rifampicin-indeterminate Xpert MTB/RIF results), only 30 submitted samples for further testing at the central laboratory (Table 1). Of these, 21 (70%) were positive for both M. tuberculosis and rifampicin resistance, and nine (30%) were positive for M. tuberculosis but indeterminate for rifampicin resistance. On repeat Xpert MTB/RIF testing at the central laboratory, of the 21 patients initially positive for rifampicin resistance, 17 (81%) were again positive for rifampicin resistance, three (14%) were negative for rifampicin resistance and one (5%) tested negative for M. tuberculosis.
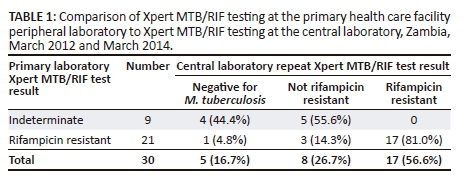
Of the nine patients initially indeterminate for rifampicin resistance, five (56%) tested negative for rifampicin resistance and four (44%) tested negative for M. tuberculosis on repeat Xpert MTB/RIF testing (Table 1). These four (44%) samples negative for M. tuberculosis on repeat Xpert MTB/RIF testing were M. tuberculosis positive but rifampicin resistance negative when tested by both MTBDRplus and MGIT drug susceptibility testing (Tables 2-3). Xpert MTB/RIF and MTBDRplus were 100% concordant for detection of rifampicin resistance (Table 2). When compared to MGIT drug susceptibility testing, the sensitivity of Xpert MTB/RIF and MTBDRplus for detection of rifampicin resistance was 100%, while specificity was 72% for Xpert MTB/RIF and 80% for MTBDRplus. Three patients had positive rifampicin resistance results on both Xpert MTB/RIF and MTBDRplus (Table 2), but rifampicin resistance was not detected with MGIT drug susceptibility testing (Tables 3 and 4). On Xpert MTB/RIF all three samples showed no signal for probe D and on MTBDRplus, all were missing the wild-type 7 probe and showed no mutation probe (results not shown). These three isolates were also resistant to isoniazid on both MGIT drug susceptibility testing and MTBDRplus (Tables 3 and 4).
The overall positive predictive values and negative predictive values for MDR tuberculosis diagnosis for Xpert MTB/RIF compared to MGIT drug susceptibility testing were 62.5% and 100% and compared to MTBDRplus were 81.2% and 100% (results not shown). For previously treated patients, the Xpert MTB/RIF positive predictive value was 77.8% and negative predictive value was 100%; for new patients, positive predictive value was 47.8% and negative predictive value was 100%. The MTBDRplus positive predictive value was 78.6% and negative predictive value was 100% compared to MGIT.
Discussion
Indeterminate rifampicin resistance results can be resolved by repeating the test, as shown in our study. Of all the samples that were initially indeterminate for rifampicin resistance, none tested positive on repeat testing. An indeterminate rifampicin resistance Xpert MTB/RIF result maybe due to low DNA quantity in the sample; this was supported by the high proportion of samples that were initially indeterminate that did not detect M. tuberculosis on repeat testing with Xpert MTB/RIF but were positive on M. tuberculosis culture. For prediction of MDR tuberculosis, our findings confirm that Xpert MTB/RIF has a low positive predictive value for MDR tuberculosis in low-prevalence MDR tuberculosis settings like Zambia, which has an estimated MDR prevalence of 4.2%.10 Depending on Xpert MTB/RIF alone for diagnosis of MDR tuberculosis in our setting would lead to subjecting a significant proportion of patients to unnecessary second-line treatment.1
The performance of Xpert MTB/RIF and MTBDRplus were similar, which was expected since both are genotypic tests. The sensitivity of Xpert MTB/RIF for detection of rifampicin resistance was within the range of what has been reported by others, whereas the specificity of 73% was lower.11 The prevalence of silent rpoB gene mutations that do not confer phenotypic resistance determines the specificity of genotypic tests, if using a phenotypic test as a gold standard. While we were limited by a lack of capacity to sequence the three samples that had results suggesting false rifampicin resistance, other studies have reported false Xpert MTB/RIF or MTBDRplus rifampicin resistance caused by silent mutations in the rpoB gene.12,13,14,15,16 Further, these three isolates were also resistant to isoniazid on both MGIT drug susceptibility testing and MTBDRplus, consistent with the widely observed fact that in most cases, silent mutations in the rpoB gene are associated with mutations in the katG gene or promoter of the inhA gene.17,18 Probe D in the Xpert MTB/RIF and the wild-type 7 probe in MTBDRplus both target the same region of the rpoB gene, codon 526, which is one of the regions with known mutations that confer resistance to rifampicin. Detection of mutations in this region by both Xpert MTB/RIF and MTBDRplus shows strong evidence of the accuracy of our genotypic results. However, MGIT drug susceptibility testing has been shown to produce false susceptibility results for strains with minimal inhibitory concentrations for rifampicin close to the cut-off value of 1 µg/mL used in MGIT drug susceptibility testing, or strains that have sub-critical minimal inhibitory concentrations for rifampicin.19 However, it is unlikely that the discordant results in our study were due to false susceptibility results for the MGIT drug susceptibility testing. To resolve these discrepant results, sequencing is required to determine the presence of silent mutations. However, lack of access to sequencing services was a limitation in our study.
This study was also limited by its small sample size but provides some insights into the comparable performance of Xpert MTB/RIF, MTBDRplus and culture drug susceptibility testing culture for the diagnosis of drug-resistant tuberculosis. There is an urgent need for Zambia to perform a full identification and classification of rpoB mutations and investigate minimal inhibitory concentrations for rifampicin, so as to optimise national guidelines for diagnosis of drug-resistant tuberculosis. A thorough investigation of the performance of Xpert MTB/RIF and MTBDRplus for diagnosis of rifampicin resistance and prediction of MDR tuberculosis in the Zambian setting is recommended to avoid inappropriate treatment. Whole genome sequencing capacity is required to fully characterise circulating rifampicin-resistant M. tuberculosis strains. Improvements are needed to make these genotypic tests function as stand-alone tests, as they offer the best prospects for early and accurate diagnosis for tuberculosis and MDR tuberculosis.
Acknowledgements
The authors would like to acknowledge the Ministry of Health of Zambia for permission to conduct the study and Lusaka District Health staff at the facilities where this study was conducted.
Competing interests
The authors declare that they have no financial or personal relationships which may have inappropriately influenced them in writing this article.
Sources of support
This study was a sub-study of the Xpert Evaluation study funded by the Stop TB Partnership/TB REACH wave 2: T9-370-114.
Authors' contributions
M.M. and H.A. conceptualised the study. D.M. and N.K. designed the databases. M.C.L., M.M.C., S.M., M.M., D.M. and W.M. performed the experiments. M.M. analysed the data. W.M. and M.M. wrote the manuscript. P.d.H. critically reviewed the manuscript. All authors reviewed and approved the final version of the manuscript.
References
1. World Health Organization. Policy statement: Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance. Geneva: WHO; 2011. [ Links ]
2. World Health Organization. Policy statement. Molecular line probe assays for rapid screening of patients at risk of multidrug resistant tuberculosis (MDR-TB) [homepage on the Internet]. [cited 2008 June 27]. Available from: http://www.who.int/tb/features_archive/policy_statement.pdf [ Links ]
3. Siu GK, Zhang Y, Lau TCK, et al. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2011;66(4):730-733. https://doi.org/10.1093/jac/dkq519 [ Links ]
4. Jamieson FB, Guthrie JL, Neemuchwala A, et al. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol. 2014;52(6):2157-2162. https://doi.org/10.1128/JCM.00691-14 [ Links ]
5. Horng YT, J W-Y, Chen Y-Y, et al. Molecular analysis of codon 548 in the rpoB gene involved in Mycobacterium tuberculosis resistance to rifampin. Antimicrob Agents Chemother. 2015;59(3):1542-1548. https://doi.org/10.1128/AAC.04374-14 [ Links ]
6. Muyoyeta M, Moyo M, Kasese N, et al. Implementation research to inform the use of Xpert MTB/RIF in primary health care facilities in high TB and HIV settings in resource constrained settings. PLoS One. 2015;10(6):e0126376. https://doi.org/10.1371/journal.pone.0126376 [ Links ]
7. Muyoyeta M, et al. Sensitivity, specificity, and reproducibility of the Capilia TB-Neo assay. J Clin Microbiol. 2013;51(12):4237-4239. https://doi.org/10.1128/JCM.02441-13 [ Links ]
8. Muyoyeta M, Mwanza WC, Kasese N, et al. Evaluation of the Capilia TB assay for culture confirmation of Mycobacterium tuberculosis infections in Zambia and South Africa. J Clin Microbiol. 2010;48(10):3773-3775. https://doi.org/10.1128/JCM.01688-09 [ Links ]
9. HAIN Life Sciences. GenoType MTBDRplus: Instructions for use. Nehren, Germany: HAIN Life Sciences. [ Links ]
10. Kapata N, Chanda-Kapata P, Bates M, et al. Multidrug-resistant TB in Zambia: Review of national data from 2000 to 2011. Trop Med Int Health. 2013;18(11):1386-1391. https://doi.org/10.1111/tmi.12183 [ Links ]
11. Steingart KR, Schiller I, Horne DJ, et al. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;1:CD009593. [ Links ]
12. Marlowe EM, Novak-Weekley SM, Cumpio J, et al. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 2011;49(4):1621-1623. https://doi.org/10.1128/JCM.02214-10 [ Links ]
13. Claessens J, Mathys V, Derdelinckx I, Saegeman V. Case report of a false positive result of the Xpert(R) MTB/RIF assay for rifampicin resistance in Mycobacterium tuberculosis complex. Acta Clin Belg. 2017 Jun;72(3):195-197. doi: 10.1179/2295333715Y.0000000072. [ Links ]
14. Mathys V, Van de Vyvere M, De Droogh E, Soetaert K, Groenen G. False-positive rifampicin resistance on Xpert(R) MTB/RIF caused by a silent mutation in the rpoB gene. Int J Tuberc Lung Dis. 2014;18(10):1255-1257. https://doi.org/10.5588/ijtld.14.0297 [ Links ]
15. Alonso M, Palacios JJ, Herrantz M, et al. Isolation of Mycobacterium tuberculosis strains with a silent mutation in rpoB leading to potential misassignment of resistance category. J Clin Microbiol. 2011;49(7):2688-2690. https://doi.org/10.1128/JCM.00659-11 [ Links ]
16. Williamson DA, Basu I, Bower J, Freeman JT, Henderson G, Roberts SA. An evaluation of the Xpert MTB/RIF assay and detection of false-positive rifampicin resistance in Mycobacterium tuberculosis. Diagn Microbiol Infect Dis. 2012;74(2):207-209. https://doi.org/10.1016/j.diagmicrobio.2012.06.013 [ Links ]
17. Ocheretina O, Byrt E, Mabou M-M, et al. False-positive rifampin resistant results with Xpert MTB/RIF version 4 assay in clinical samples with a low bacterial load. Diagn Microbiol Infect Dis. 2016;85(1):53-55. https://doi.org/10.1016/j.diagmicrobio.2016.01.009 [ Links ]
18. N'Guessan K, Assi JS, Ouassa T, et al. Assessment of the genotype MTBDRplus assay for rifampin and isoniazid resistance detection on sputum samples in Cote d'Ivoire. Eur J Microbiol Immunol (Bp). 2014;4(3):166-173. https://doi.org/10.1556/EUJMI-D-14-00014 [ Links ]
19. Ocheretina O, Escuyer VE, Mabou M-M, et al. Correlation between genotypic and phenotypic testing for resistance to rifampin in Mycobacterium tuberculosis clinical isolates in Haiti: Investigation of cases with discrepant susceptibility results. PLoS One. 2014;9(3):e90569. https://doi.org/10.1371/journal.pone.0090569 [ Links ]
 Correspondence:
Correspondence:
Winnie Mwanza
winnie_mwanza@yahoo.co.uk
Received: 09 Apr. 2018
Accepted: 26 Sept. 2018
Published: 06 Dec. 2018














