Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Enology and Viticulture
versión On-line ISSN 2224-7904
versión impresa ISSN 0253-939X
S. Afr. J. Enol. Vitic. vol.42 no.2 Stellenbosch 2021
http://dx.doi.org/10.21548/42-2-4724
ARTICLES
A Shotgun Metagenomic Sequencing Exploration of Cabernet Sauvignon Grape Must Reveals Yeast Hydrolytic Enzymes
S. Ghosh†, *; B. Divol; M.E. Setati*
South African Grape and Wine Research Institute, Stellenbosch University, Stellenbosch, South Africa
ABSTRACT
Shotgun sequencing was employed to explore the community structure (phylotyping of rRNA genes) and functional potential of Cabernet Sauvignon grape must microbiome. A metagenomic library, representing 92.6 Mb of genetic information, was generated from DNA obtained from Cabernet Sauvignon grape must.. Fungi were identified as the dominant domain (59.5%) followed by Streptophyta (39%). Among the 84 fungal species, 22 were yeasts of various genera. Additionally, grapevine endophytes such as Davidiella sp., Botryotinia fuckeliana, Alternaria sp., and Cladosporium sp. were identified. An unusually high prevalence of Mucor spp. was evidenced. Functional annotation revealed sequences of genes involved in metabolism (35.6%), followed by poorly characterized categories (28.3%), cellular processes and signalling (18.4%), and finally information storage (17.8%). Among the former, glycosidases were abundant followed by glycogen debranching enzyme, 6-phosphofructokinase and trehalose-6-phosphate synthase. Furthermore, the taxonomic analysis of the functional sequence data exhibited the eukaryotic gene pool that predominantly contains sequences derived from Streptophyta (mainly Vitis vinifera) 60% > Ascomycota (32%) > Basidiomycota (5%) > Bacteria (2.5%). Finally, sequences of a variety of hydrolytic enzymes of potential oenological relevance were retrieved, thereby confirming that grape juice is a rich reservoir for valuable biocatalysts that should be explored further.
Key words: Shotgun sequencing, metagenomic DNA, grape juice, microbiota, hydrolytic enzymes
INTRODUCTION
The grape and wine ecosystem is a complex environment that encompasses different species of filamentous fungi, yeasts and bacteria The epiphytic filamentous fungi mainly comprise species of the genera Aspergillus, Botrytis, Alternaria, Penicillium and Cladosporium (Diguta et al., 2011) while the endophytes consist of Alternaria spp., Epicoccum nigrum, Leptosphaerulina chartarum, Aureobasidium pullulans, Botryosphaeria spp. amongst other species (Pancher et al., 2012). In the vineyard, the occurrence frequency and intensity of these populations depend on the developmental stages of the grape. Indeed, it has been reported that the frequency of filamentous fungi varies from 104 - 106 cfu/g berry (Da et al., 2002;Diguta et al., 2011). The yeast population comprises both basidiomycetous and ascomycetous species. The basidiomycetous yeasts, such as Cryptococcus spp., Rhodotorula spp., Sporobolomyces spp. and the yeastlike ascomycetous fungus A. pullulans, are predominantly present on intact unripe grape berries. In contrast, ascomycetous yeasts such as Candida spp., Hanseniaspora spp., Metschnikowia spp. and Pichia spp. mostly occur on ripe berries. Both highly, Zygosaccharomyces rouxii, and weakly fermentative yeast such as Zygoascus hellenicus, is also occurred (Fleet, 2003;Loureiro and Malfeito-Ferreira, 2003;Barata et al., 2008;Barata et al., 2012b;Bokulich et al., 2016). Surprisingly, the principal wine yeast, Saccharomyces cerevisiae is rarely detected on undamaged grapes (Barata et al., 2012b). The unripe berries typically harbour a yeast population of 103 cfu/g berry while ripe berries may contain 104-106 cfu/g berry (Parish and Caroll, 1985;Fleet and Heard, 1993;Barata et al., 2012a;b). This population may increase up to 108 cfu/g berry on damaged grapes (Fleet, 2002;Barata et al., 2012b).
The most predominant bacterial populations are lactic acid bacteria (LAB) and acetic acid bacteria (AAB). Many LAB such as Lentilactobacillus hilgardii (formerly Lactobacillus hilgardii, Lactiplantibacillus plantarum (formerly Lactobacillus plantarum), Lacticaseibacillus casei (formerly Lactobacillus casei), Oenococcus oeni, Leuconostoc mesenteroides, Pediococcus damnosus and Pediococcus parvulus have been isolated from grape surfaces (Renouf et al., 2008;Barata et al., 2012b;Martins et al., 2012;Salvetti et al., 2016). Few acetic acid bacteria, e.g. Gluconobacter spp., Acetobacter spp. have also been reported from intact as well as damaged grapes at 102103 cfu/g and 106- 108 cfu/g berries, respectively (Barata et al., 2012a). Other bacterial species such as Enterobacter spp., Bacillus spp., Burkholderia spp., Serratia spp. and Staphylococcus spp. have also been occasionally reported to be present on the surface of grape berries in lower levels (Barata et al., 2012b).
Upon crushing of the grape bunches, all the microorganisms described above end in the grape must to various degrees, depending on their ability to survive the environmental conditions occurring in grape must. Ultimately, these microorganisms contribute to the final composition of wine through their biochemical activities that include their extracellular hydrolytic enzyme activities. Although filamentous fungi and the yeast-like fungus A. pullulans do not participate in the wine fermentation, they have been reported to possess enzymes of oenological interest such as pectinases and glucanases (van Rensburg and Pretorius, 2000;Bozoudi and Tsaltas, 2018). Among the yeasts, the non-Saccharomyces species produce an array of extracellular hydrolytic enzymes as previously reviewed (Jolly et al., 2014;García et al., 2016;Mateo and Maicas, 2016;Padilla et al., 2016). For instance, Hanseniaspora spp., Debaryomyces spp., Candida spp., Pichia spp. and Torulaspora spp., Hansenula sp., have been reported to produce extracellular hydrolytic enzymes like glucosidases, pectinases and proteases (Charoenchai et al., 1997;Strauss et al., 2001;Bueso et al., 2012;Maturano et al., 2012;García et al., 2016;Mateo andMaicas, 2016;Padilla et al., 2016). These extracellular enzymes catalyse different types of reactions in must and during fermentation. For instance, glycosidases hydrolyse the non-volatile precursors from grapes releasing certain volatile compounds (e.g. monoterpenes), thereby improving the wine flavour and aroma (Charoenchai et al., 1997). Pectinases (polygalacturonases, pectin lyase, pectinesterase, acetylesterase) facilitate the juice extraction from grapes by lowering the viscosity of the grape juice, improving wine clarification and facilitating filtration (van Rensburg and Pretorius, 2000;Claus and Mojsov, 2018b). Some of the non-Saccharomyces yeast species are reported to exhibit proteolytic activities. The enzymes catalyzing these activities are mainly involved in hydrolysis of proteins and they have been proposed as additives to prevent protein haze formation, although their application is not yet effective (Lagace and Bisson, 1990;Pocock et al., 2003;Jolly et al., 2006).
Most studies of these enzymes have been conducted on the individual microbial isolates (Charoenchai et al., 1997;Strauss et al., 2001). However, these approaches only allow access to enzymes from a few selected organisms derived from the culturable microbiota (Lorenz et al., 2002) thus making metagenomic approaches more desirable to increase the chances of retrieving enzymes derived from other microorganisms that might be excluded through culture-dependent methods. Metagenomic techniques have been implemented and indeed proved successful in retrieving biotechnologically relevant microbial enzymes including chitinases, dehydrogenases, proteases, oxygenases, lipases, nitralases, esterases, amylases, xylanase and polyketide synthases both from extreme and non-extreme environments (Yeh et al., 2013;Schroder et al., 2014). However, these approaches have never been employed to mine the wine microbiome.
The focus of the present exploratory study was to explore of the grape juice microbiome and its functional potential with a specific focus on hydrolytic enzymes of oenological interest. Direct DNA shotgun sequencing and sequence-based screening for enzymes of oenological relevance were performed.
MATERIALS AND METHODS
Metagenomic Sample collection and DNA extraction
Cabernet Sauvignon grapes (5 kg) were collected from a biodynamic vineyard (33°57'39.33" S 18° 45'13.46" E elev 183 m) located in the Stellenbosch wine producing region of South Africa. The grapes were hand destemmed and crushed in a pre-sterilized beaker. A 50 mL sample of the fresh must was collected and centrifuged at 5000 rpm for 10 min. The pellet was washed three times with EDTA-PVP solution containing 0.15 M NaCl, 0.1 M EDTA and 2% (w/v) Polyvinylpyrolidone (Jara et al., 2008) followed by three washes with TE buffer (pH 7.6). DNA extraction was carried out according to (Wilson, 1997) with minor modifications as follows. The pellet was re-suspended in 2.3 mL TE buffer, followed by the addition of proteinase K, SDS and 500 uL of fine glass beads. The mixture was vortexed for 3 min. Twenty microlitres of a 10 mg/mL lysozyme solution, were added and the mixture incubated at 37°C for 50 min. Thereafter, 400 uL of 5 M NaCl and 240 uL CTAB (2%) (5M) /NaCl solutions were added, and the mixture was incubated for 10 min at 65°C, followed by phenol/chloroform/isoamyl alcohol extraction and precipitation with isopropanol as described in Wilson (2001).
Pyrosequencing and sequence data quality control
Approximately 500 ng of genomic DNA with a 260/280 ratio > 1.8 was used as a template for shotgun pyrosequencing on a Genome Sequencer (GS) FLX system (Roche Applied Science, Manheim, Germany) using Titanium chemistry, which was performed by Inqaba Biotec (Pretoria, South Africa). Two independent libraries were prepared. The DNA libraries were constructed according to the GS FLX Rapid Library Preparation kit (Roche Applied Science) and the optimal DNA copy per bead ratio was determined by emulsion PCR titration using the GS FLX Titanium SV emPCR kit (Lib-L; Roche Applied Science). The final emulsion PCR for sequencing production was performed suing the GS FLX Titanium LV emPCR kit (Lib-L, Roche Applied Science).
Data analysis
Raw 454 sff files were uploaded on the MetaGenome Rapid Annotation Subsystems Technology (MG-RAST) server (Meyer et al., 2008). The shotgun sequencing reads are available to the public on MG-RAST under the project Bio_IWBT1 with the following IDs mgm4512887.3 and mgm4519241.3 (https://www.mg-rast.org/linkin.cgi?project=mgp3298). The data underwent quality control (QC) analyses including quality filtered, length filtered, and dereplication. Taxonomic analysis was done by comparison of the metagenome data with the M5RNA database available on MG-RAST using a minimum identity cut-off of 97% and a minimum alignment length of 150 bp. To characterize the gene content of the grape must, all reads were functionally annotated by means of the Clusters of Orthologous Groups of proteins database (COG) (Tatusov et al., 2001;Tatusova et al., 2014). Organism and functional identifications were performed using a BLAT [Basic Local Alignment Search Tool (BLAST)-like alignment tool] search of the integrative MG-RAST M5NR database, which is a non-redundant protein database that combines sequences from multiple common sources. Identifications were made using a maximum e-value of 1e-8, a minimum identity cut-off of 60% and a minimum alignment length of 50 amino acids. The relative abundance of each gene or species was determined by dividing the number of hits of that particular gene or species by the total number of hits.
RESULTS
A metagenomic library was generated from total DNA extracted from a Cabernet Sauvignon grape must. Two sequencing runs (datasets A and B) generated a total of 175,616 reads accounting for 92,570,157 bps of sequence information (Table 1). A total of 148,845 reads were designated as high-quality sequences post QC.
Analysis of grape must-associated microbiota
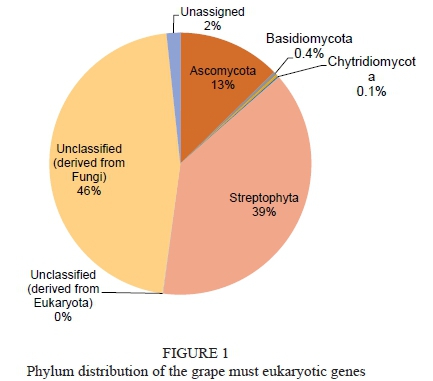
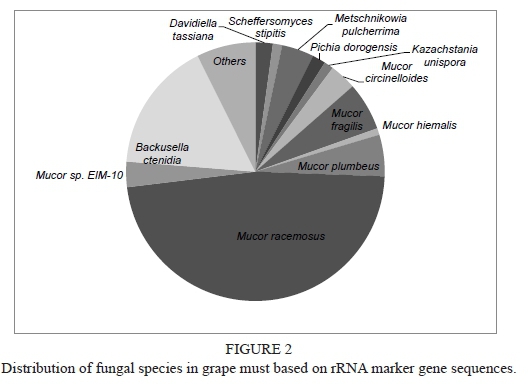
Only 2.59% of the total sequence data represented rRNA genes, mostly distributed within the SILVA (Latin silva, forest, http://www.arb-silva.de)-LSU (Large subunit ribosomal) and SSU (Small subunit ribosomal) databases. The MG-RAST classification tool revealed that eukaryotes were the dominant domain (Fig. 1) with Streptophyta accounting for 39% of the sequences and fungi for 59.5%. Further analysis of the fungal composition revealed a high diversity with 84 fungal species comprising 22 yeast species including known wine yeasts such as members of the genera Hanseniaspora, Candida, Lachancea, Metschnikowia, Pichia, Torulaspora, Saccharomyces, Zygosaccharomyces, Saccharomycopsis and Kluyveromyces (Fig. 2). In addition, typical grapevine endophytes including Davidiella sp., Botryotinia fuckeliana, Alternaria sp., as well as Cladosporium sp. were also detected. Notably, it has been observed that there is an incidence of Mucor spp.
Functional analysis
To gain insight into the biological processes represented in the wine metagenome, sequences were annotated according to Clusters of Orthologous Groups (COG) of proteins categories thus assigning predicted functions to coding sequences. As depicted in Fig. 3 genes related to metabolism accounted for the majority of the sequences (35.6%), followed by poorly characterized categories (28.3%), cellular processes and signalling (18.4%), and information storage (17.8%). Within the metabolism category, genes encoding proteins required for energy production and conversion were most abundant. Specifically, genes encoding glycosidases were the most abundant within the carbohydrate transport and metabolism gene pool, followed by glycogen debranching enzyme, 6-phosphofructokinase and trehalose-6-phosphate synthase encoding genes (Table 2). In addition, genes involved in the Calvin-Benson cycle, nitrogen and sulphur metabolism were also abundant. Degradation of benzoate, toluene and polycyclic aromatic hydrocarbon were the most represented processes in the secondary metabolic processes (Table 2).
Taxonomic analysis of the functional gene sequences showed that 60% of the eukaryotic gene pool was derived from Streptophyta (mainly Vitis vinifera), 32% from the Ascomycota, 5% from the Basidiomycota (Fig. 4A). The bacterial genes accounted for 2.5% of the sequence data with Proteobacteria being the most predominant phylum in the bacterial domain, accounting for 62% of the total sequences, followed by the Firmicutes, Bacteroidetes and Actinobacteria, which accounted for 12%, 11% and 10%, respectively (Fig. 4B).
Furthermore, the potential fungal hydrolases were identified from the carbohydrate transport and metabolism gene pool with fragment's length ranging from 400 - 600 bp (Table 3). Notably, most of the fragments were observed to be derived from the close filamentous fungal homologues (Table 3).
DISCUSSION
The winemaking process is primarily governed by an array of enzymatic activities that drive the fermentation kinetics involved in the bioconversion of grape juice to wine. These enzymes are derived from grapes and the microbiota present on/in the grape/must/wine (van Rensburg and Pretorius, 2000). The current study evaluated the functional potential of the grape must metagenome. Unsurprisingly, the data show that Vitis vinifera is the dominant organism detected within the grape must ecosystem followed by fungi of the phylum Ascomycota. Further investigations of the fungal community confirmed the presence of a variety of filamentous fungi, the yeast-like fungus Aureobasidium pullulans and several non-Saccharomyces yeast genera such as Clavispora, Metschnikowia, Kazachstania, Torulaspora, Lachancea, Candida, Pichia and Kluyveromyces. Members of these genera are also often frequently retrieved from grape must either through culture-based methods, DGGE as well as amplicon sequencing (Bokulich et al., 2014;David et al., 2014;Taylor et al., 2014;Pinto et al., 2015;Setati et al., 2015;Wang et al., 2015;Kecskeméti et al., 2016;De Filippis et al., 2017). Analysis of the functional genes revealed that Streptophyta (60%) followed by Ascomycota (32%) and Basidiomycota (5%) contributed the most to the metagenome. Furthermore, the COG based analysis of the coding sequences showed homology to an array of genes encoding hydrolytic enzymes (carbohydrate metabolism). Such observations were reported previously with genes related to amino acid metabolism, transport, transcription and carbohydrate metabolism (Campanaro et al., 2014;Salvetti et al., 2016). As depicted Table 2 and Table 3, in the carbohydrate transport and metabolism gene pool, glycosidases (26 hits) were the dominant enzymes followed by chitinases, endoglucanases, glycogen debranching enzyme and 6-phosphofructokinase encoding genes. The fungal partial sequences of the genes encoding putative hydrolases were found to originate from filamentous fungi and non-Saccharomyces yeasts (Table 3), with multiple sequences matching oenologically relevant enzymes from different genera (e.g. glucanases) or from the same genera (e.g. different glucosidases from Talaromyces) or from the same species (e.g. an aspartic protease and chitinase from Metschnikowia pulcherrima). ß-1,3-Exoglucanases such as that of Ampelomyces quisqualis and Trichoderma spp., to which sequence ID: H551VLF01BWCP7 and ID: H551VLF01A5JYC exhibited homology, respectively, have been implicated in mycoparasitic interactions (Rotem et al., 1999) and are exploited in biocontrol against grapevine powdery mildew disease (Falk et al., 1995) and other diseases (Cabaroglu et al., 2003;Punja and Utkhede, 2003;Sanz et al., 2005). Our metagenomic sequences also revealed a few sequences that showed close homology to ß-glucosidase genes from Aspergillus spp. and Talaromyces spp. Aspergillus spp. has been previously reported as potential biocontrol agent (Tiwari et al., 2011;Rosada et al., 2013) and also shown to secrete ß-glucosidase (Watanabe et al., 1992;Vaithanomsat et al., 2011) that are beneficial for the wine production (Maicas, 2016). Talaromyces sp. is not a frequent fungus isolated from grape. It has however been reported that this fungus exhibits glucosidase activity (Nakkharat and Haltrich, 2006). Only a few sequences of genes encoding enzymes involved in protein degradation were retrieved. Interestingly, one sequence ID: HQ7JFPR01BHOWG exhibited 52% identity to the aspartic protease from M. pulcherrima (Reid et al., 2012). Addition of enzymes derived from grape-associated fungi during grape juice and wine processing, has become common practice since these enzymes offer benefits such as increased juice yield, improved colour extraction, clarification and accelerated settling (Manzanares et al., 1999;Fernandez et al., 2000;Belda et al., 2016;Claus and Mojsov, 2018a). In particular, aspartic proteases have been proposed as potential alternative to other wine stabilisation methods such as bentonite fining (Theron and Divol, 2014).
The functional sequence profile of the prokaryotic population in our study was mainly dominated by Proteobacteria, followed by Firmicutes, Bacteroidetes and Actinobacteria (Fig. 4B). Earlier, high throughput analysis of the grape berry surface, phyllosphere and flowers has also reported such findings (Perazzolli et al., 2014;Pinto et al., 2014;Pinto et al., 2015;Portillo et al., 2016;Portillo and Mas, 2016). Furthermore, a study by Salvetti el al., 2016 which was conducted on both traditional (TW) and accelerated (AW) withering conditions berries using high throughput shot gun sequencing has also reported the presence of the Proteobacteria as the predominant phylum with 97.7% and 86.1% from both TW and AW berries respectively followed by Firmicutes and Bacteroidetes (Salvetti et al., 2016). However, a former study reported that, as the fermentation progresses, the Proteobacteria population declines and that of the Firmicutes increases (Bokulich et al., 2012). When the genetic information were further analysed for genes encoding bacterial proteins/enzymes, the Enterobacter and Bacteroidetes were found to be the predominant ones; however, their exact role in wine has not been elucidated even though they are frequently encountered in grape must (Bokulich et al., 2012;Pinto et al.,2015; Portillo et al., 202016;Salvetti et al., 2016). Previous studies have shown that the enzymes, such as glucosidases (Davis et al., 1985) and glycosidases (Grimaldi et al., 2000) secreted from Oenococcus oeni contributed largely to the floral and fruity characteristics of wine by releasing monoterpenes, norisoprenoids and aliphatic compounds. Other bacteria such as Leuconostoc, Lactobacillus and Pediococcus have also been reported to contribute to wine final aroma (Williams et al., 1982;Ugliano et al., 2003;D'Incecco et al., 2004). The whole metagenomic sequencing is a valuable technique to reveal biocatalysts that have not yet been retrieved from cultured microorganisms. However, the sequence data obtained from shotgun sequences only provided a snapshot of the functional potential of the wine microbiota since only partial gene sequences were obtained and the sequence coverage/depth was not enough to provide a full representation of the entire metagenome. Indeed, only 92.5 Mb of sequence data were retrieved, representing approximately 7 yeast genome sizes which could be improved if better high throughput sequencing techniques and/or platforms are employed. Nevertheless, the data showed that glucosidases are the most predominant glycosyl hydrolases in the grape must ecosystem. Indeed, most phenotypical screening attempts have similar trends even though the focus tends to be on non-Saccharomyces yeasts (Charoenchai et al., 1997;van Rensburg and Pretorius, 2000;Strauss et al., 2001;Maturano et al., 2012). Moreover, the glycosyl hydrolase-encoding genes showed a percentage similarity ranging from 42 to 88% to known fungal species (Table 3). This finding was not surprising because the lack of fully annotated non-Saccharomyces genomes poses a challenge in gene prediction and annotation of the metagenomes. Many of the genomes of wine related non-Saccharomyces yeasts are still not sequenced and those which are not fully annotated . In conclusion, our study intended to explore if DNA shotgun sequencing could provide a wide, unbiased view of the wine microbial diversity, encompassing both the taxonomical and functional potential, in a single snapshot with high resolution. To the best of our knowledge, this first metagenomic study has accounted for the functional aspects of the grape must microbiome. The study confirms that grape juice is a rich reservoir ofvaluable biocatalysts. Indeed, hydrolytic enzymes from oenological relevance were successfully retrieved from direct sequencing data sets. Further investigations should include retrieving full gene sequences and thereafter test their expression during the wine fermentation. The activity of the corresponding enzymes under winemaking conditions should also be assessed in order to evaluate their impact of their activities on the sensory properties of wine.
LITERATURE CITED
Barata, A., Gonzalez, S., Malfeito-Ferreira, M., Querol, A. and Loureiro, V., 2008. Sour rot-damaged grapes are sources of wine spoilage yeasts. FEMS Yeast Res. 8, 7, 1008-1017. [ Links ]
Barata, A., Malfeito-Ferreira, M. and Loureiro, V., 2012a. Changes in sour rotten grape berry microbiota during ripening and wine fermentation. Int. J. Food Microbiol. 154, 3, 152-161. [ Links ]
Barata, A., Malfeito-Ferreira, M. and Loureiro, V., 2012b. The microbial ecology of wine grape berries. Int. J. Food Microbiol., 153, 3, 243-259. [ Links ]
Belda, I., Ruiz, J., Alastruey-Izquierdo, A., Navascues, E., Marquina, D. and Santos, A., 2016. Unraveling the Enzymatic Basis of Wine "Flavorome": A Phylo-Functional Study of Wine Related Yeast Species. Front. Microbiol., 7, 12. [ Links ]
Bokulich, N.A., Collins, T.S., Masarweh, C., Allen, G., Heymann, H., Ebeler, S.E. and Mills, D.A., 2016. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. Mbio, 7, 3, e00631-16 . [ Links ]
Bokulich, N.A., Joseph, C.M., Allen, G., Benson, A.K. and Mills, D.A., 2012. Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS One 7, e36357. [ Links ]
Bokulich, N.A., Thorngate, J.H., Richardson, P.M. and Mills, D.A., 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, 1, E139-148. [ Links ]
Bozoudi, D. and Tsaltas, D., 2018. The multiple and versatile roles of Au-reobasidium pullulans in the vitivinicultural sector. Fermentation 4, 4, 85. [ Links ]
Bueso, C.G., Zarzoso, B.E., Cabellos, J.M., Díaz, G.M. and Arroyo, T., 2012. Biotechnological potential of non-Saccharomyces yeasts during isolated during spontaneous fermentations of Malvar (Vitis vinifera cv. L.). Eur. Food Res. Technol., 236, 193-207. [ Links ]
Cabaroglu, T., Selli, S., Canbas, A., Lepoutre, J.P. and Günata, Z., 2003. Wine flavor enhancement through the use of exogenous fungal glycosidases. Enzyme Microbial Technol. 33, 5, 581-587. [ Links ]
Campanaro, S., Treu, L., Vendramin, V., Bovo, B., Giacomini, A. and Co-rich, V., 2014. Metagenomic analysis of the microbial community in fermented grape marc reveals that Lactobacillus fabifermentans is one of the dominant species: insights into its genome structure. Appl. Microbiol. Bio-technol. 98, 13, 6015-6037. [ Links ]
Charoenchai, C., Fleet, G.H., Henschke, P.A. and Todd, B.E.N., 1997. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 3, 1, 2-8. [ Links ]
Claus, H. and Mojsov, K., 2018. Enzymes for wine fermentation: current and perspective applications. Fermentation 4, 3, 52. [ Links ]
D'incecco, N., Bartowsky, E., Kassara, S., Lante, A., Spettoli, P. and Henschke, P., 2004. Release of glycosidically bound flavour compounds of Chardonnay by Oenococcus oeni during malolactic fermentation. Food Microbiol. 21 , 3, 257-266. [ Links ]
Da, R.R., Palacios, V., Combina, M., Fraga, M.E., De, O.R., Magnoli, C.E. and Dalcero, A.M., 2002. Potential ochratoxin A producers from wine grapes in Argentina and Brazil. Food Addit. Contam. 19, 4, 408-414. [ Links ]
David, V., Terrat, S., Herzine, K., Claisse, O., Rousseaux, S., Tourdot-Marechal, R., Masneuf-Pomarede, I., Ranjard, L. and Alexandre, H., 2014. High-throughput sequencing of amplicons for monitoring yeast biodiversity in must and during alcoholic fermentation. J. Ind. Microbiol. Biotechnol. 41, 5, 811-821. [ Links ]
Davis, C.R., Wibowo, D., Eschenbruch, R., Lee, T.H. and Fleet, G.H., 1985. Practical implications of malolactic fermentation, a review. Am. J. Enol. Vitic. 36, 290-301. [ Links ]
De Filippis, F., La Storia, A. and Blaiotta, G., 2017. Monitoring the myco-biota during Greco di Tufo and Aglianico wine fermentation by 18S rRNA gene sequencing. Food Microbiol. 63, 117-122. [ Links ]
Diguta, C.F., Vincent, B., Guilloux-Benatier, M., Alexandre, H. and Rous-seaux, S., 2011. PCR ITS-RFLP: A useful method for identifying filamentous fungi isolates on grapes. Food Microbiol. 28, 6, 1145-1154. [ Links ]
Falk, S.P., Gadoury, D.M., Pearson, R.C. and Seem, R.C., 1995. Partial Central of Grape Powdery Mildew by the Mycoparasite Ampelomyces-Quisqua-lis. Plant Dis. 79, 5, 483-490. [ Links ]
Fernandez, M., Ubeda, J.F. and Briones, A.I., 2000. Typing of non-Saccha-romyces yeasts with enzymatic activities of interest in wine-making. Int. J. Food Microbiol. 59, 1-2, 29-36. [ Links ]
Fleet, G.H., 2003. Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 1-2, 11-22. [ Links ]
Fleet, G.H. and Heard, G.M., 1993. Yeast growth during fermentation. In: Fleet, G.H. (ed). Wine Microbiology and Biotechnology, Harwood Academic, Lausanne, pp. 27-54. [ Links ]
Fleet, G.H., Prakittchaiwattana, C., Beh, A.L. and Heard, G.M., 2002. The yeast ecology of wine grapes. In: Ciani, M. (ed). Biodiversity and biotechnology of wine yeasts, Research Signpost, Kerala, pp. 1-17. [ Links ]
García, M., Esteve-Zarzoso, B. and Arroyo,, T. 2016. Non-Saccharomyces yeasts: biotechnological role for wine production. In: Morata, A. and Loira, I. (eds). Grape and Wine Biotechnology. IntechOpen, DOI: 10.5772/64957 [ Links ]
Grimaldi, A., Mclean, H. and Jiranek, V., 2000. Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacterium, Oenococcus oeni. Am. J. Enol. Vitic. 51, 4, 362-369. [ Links ]
Jara, C., Mateo, E., Guillamon, J.M., Torija, M.J. and Mas, A., 2008. Analysis of several methods for the extraction of high quality DNA from acetic acid bacteria in wine and vinegar for characterization by PCR-based methods. Int. J. Food Microbiol. 128, 2, 336-341. [ Links ]
Jolly, N., Augustyn, O. and Pretorius, I.S., 2006. The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 27, 1, 1539. [ Links ]
Jolly, N.P., Varela, C. and Pretorius, I.S., 2014. Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 2, 215-237. [ Links ]
Kecskeméti, E., Berkelmann-Löhnertz, B. and Reineke, A., 2016. Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS One 11, 8, e0160852. [ Links ]
Lagace, L.S. and Bisson, L.F., 1990. Survey of yeast acid proteases for effectiveness of wine haze reduction. Am. J. Enol. Vitic. 41 , 2, 147-155. [ Links ]
Lorenz, P., Liebeton, K., Niehaus, F. and Eck, J., 2002. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13, 6, 572-577. [ Links ]
Loureiro, V. and Malfeito-Ferreira, M., 2003. Spoilage yeasts in the wine industry. Int. J. Food Microbiol. 86, 1-2, 23-50. [ Links ]
Manzanares, P., Ramon, D. and Querol, A., 1999. Screening of non-Saccha-romyces wine yeasts for the production of beta-D-xylosidase activity. Int. J. Food Microbiol. 46, 2, 105-112. [ Links ]
Martins, G., Miot-Sertier, C., Lauga, B., Claisse, O., Lonvaud-Funel, A., Soulas, G. and Masneuf-Pomarede, I., 2012. Grape berry bacterial micro-biota: impact of the ripening process and the farming system. Int. J. Food Microbiol. 158, 2, 93-100. [ Links ]
Maicas, S. and Mateo, J.J., 2016. Microbial Glycosidases for Wine Production. Beverages. 2, 3, 1-11. [ Links ]
Mateo, J.J. and Maicas, S., 2016. Application of non-Saccharomyces yeasts to wine-making process. Fermentation 2, 3, 14. [ Links ]
Maturano, Y.P., Rodriguez Assaf, L.A., Toro, M.E., Nally, M.C., Vallejo, M., Castellanos De Figueroa, L.I., Combina, M. and Vazquez, F., 2012. Multi-enzyme production by pure and mixed cultures of Saccharomyces and non-Saccharomyces yeasts during wine fermentation. Int. J. Food Microbiol. 155, 1-2, 43-50. [ Links ]
Meyer, F., Paarmann, D., D'souza, M., Olson, R., Glass, E.M., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J. and Edwards, R.A., 2008. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386. [ Links ]
Nakkharat, P. and Haltrich, D., 2006. Purification and characterisation of an intracellular enzyme with beta-glucosidase and beta-galactosidase activity from the thermophilic fungus Talaromyces thermophilus CBS 236.58. J. Biotechnol. 123, 3, 304-313. [ Links ]
Padilla, B., Gil, J.V. and Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7, 411. [ Links ]
Pancher, M., Ceol, M., Corneo, P.E., Longa, C.M., Yousaf, S., Pertot, I. and Campisano, A., 2012. Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Appl. Environ. Microbiol. 78, 12, 4308-4317. [ Links ]
Parish, M.E. and Caroll, D.E., 1985. Indigenous yeasts associated with Muscadine (Vitis routundifolia) grapes and must. Am. J. Enol. Vitic. 36, 2, 165-169. [ Links ]
Perazzolli, M., Antonielli, L., Storari, M., Puopolo, G., Pancher, M., Giovannini, O., Pindo, M. and Pertot, I., 2014. Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl. Environ. Microbiol. 80, 12, 3585-3596. [ Links ]
Pinto, C., Pinho, D., Cardoso, R., Custodio, V., Fernandes, J., Sousa, S., Pinheiro, M., Egas, C. and Gomes, A.C., 2015. Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6, 905. [ Links ]
Pinto, C., Pinho, D., Sousa, S., Pinheiro, M., Egas, C. and A, C.G., 2014. Unravelling the diversity of grapevine microbiome. PLoS One 9, 1, e85622. [ Links ]
Pocock, K.F., Hoj, P.B., Adams, K.S., Kwiatkowski, M.J. and Waters, E.J., 2003. Combined heat and proteolytic enzyme treatment of white wines reduces haze forming protein content without detrimental effect. Aus. J. Grape Wine Res. 9, 1, 56-63. [ Links ]
Portillo, M.C., Franquès, J., Araque, I., Reguant, C. and Bordons, A., 2016. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 219, 56-63. [ Links ]
Portillo, M.C. and Mas, A., 2016. Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding. LWT Food Sci. Technol. 72, 317-321. [ Links ]
Punja, Z.K. and Utkhede, R.S., 2003. Using fungi and yeasts to manage vegetable crop diseases. Trends Biotechnol. 21 , 9, 400-407. [ Links ]
Reid, V.J., Theron, L.W., Du Toit, M. and Divol, B., 2012. Identification and partial characterization of extracellular aspartic protease genes from Metschnikowia pulcherrima IWBT Y1123 and Candida apicola IWBT Y1384. Appl. Environ. Microbiol. 78, 19, 6838-6849. [ Links ]
Renouf, V., Claisse, O. and Lonvaud-Funel, A., 2008. Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Aus. J. Grape Wine Res. 11, 3, 316-327. [ Links ]
Rosada, L.J., Sant'anna, J.R., Franco, C.C., Esquissato, G.N., Santos, P.A., Yajima, J.P., Ferreira, F.D., Machinski, M., Correa, B. and Castro-Prado, M.A., 2013. Identification of Aspergillus flavus isolates as potential biocontrol agents of aflatoxin contamination in crops. J. Food Prot. 76, 6, 10511055. [ Links ]
Rotem, Y., Yarden, O. and Sztejnberg, A., 1999. The mycoparasite Ampelo-myces quisqualis expresses exgA encoding an exo-beta-1,3-glucanase in culture and during mycoparasitism. Phytopathology 89, 8, 631-638. [ Links ]
Salvetti, E., Campanaro, S., Campedelli, I., Fracchetti, F., Gobbi, A., Torni-elli, G.B., Torriani, S. and Felis, G.E., 2016. Whole-metagenome-sequenc-ing-based community profiles of Vitis vinifera L. cv. Corvina berries withered in two post-harvest conditions. Front Microbiol 7, 937. [ Links ]
Sanz, L., Montero, M., Redondo, J., Llobell, A. and Monte, E., 2005. Expression of an alpha-1,3-glucanase during mycoparasitic interaction of Trichoderma asperellum. FEBS J. 272, 2, 493-499. [ Links ]
Schroder, C., Elleuche, S., Blank, S. and Antranikian, G., 2014. Characterization of a heat-active archaeal beta-glucosidase from a hydrothermal spring metagenome. Enzyme Microb. Technol. 57, 48-54. [ Links ]
Setati, M.E., Jacobson, D. and Bauer, F.F., 2015. Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front Microbiol. 6, 1358. [ Links ]
Strauss, M.L.A., Jolly, N.P., Lambrechts, M.G. and Van Rensburg, P., 2001. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 91 , 1, 182-190. [ Links ]
Tatusov, R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D. and Koonin, E.V., 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 1, 22-28. [ Links ]
Tatusova, T., Ciufo, S., Fedorov, B., O'neill, K. and Tolstoy, I., 2014. Ref-Seq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 42, D553-559. [ Links ]
Taylor, M.W., Tsai, P., Anfang, N., Ross, H.A. and Goddard, M.R., 2014. Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 9, 2848-2858. [ Links ]
Theron, L.W. and Divol, B., 2014. Microbial aspartic proteases: current and potential applications in industry. Appl. Microbiol. Biotechnol. 98, 88538868. [ Links ]
Tiwari, C.K., Parihar, J. and Verma, R.K., 2011. Potential of Aspergillus niger and Trichoderma viridae as biocontrol agents of wood decay. J. Indian Acad. Wood Sci. 8, 169-172. [ Links ]
Ugliano, M., Genovese, A. and Moio, L., 2003. Hydrolysis of wine aroma precursors during malolactic fermentation with four commercial starter cultures of Oenococcus oeni. J. Agric. Food Chem. 51 , 17, 5073-5078. [ Links ]
Vaithanomsat, P., Songpim, M., Malapant, T., Kosugi, A., Thanapase, W. and Mori, Y., 2011. Production of beta-glucosidase from a newly isolated Aspergillus species using response surface methodology. Int. J. Microbiol. 2011, 949252. [ Links ]
Van Rensburg, P. and Pretorius, I.S., 2000. Enzymes in wine making: harnessing natural catalysts for efficient biotransformations - a review. S. Afr. J. Enol. Vitic. 21, 1, 52-73. [ Links ]
Wang, C., Garcia-Fernández, D., Mas, A. and Esteve-Zarzoso, B., 2015. Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front Microbiol. 6, 1-8. [ Links ]
Watanabe, T., Sato, T., Yoshioka, S., Koshijima, T. and Kuwahara, M., 1992. Purification and properties of Aspergillus niger beta-glucosidase. Eur. J. Biochem. 209, 2, 651-659. [ Links ]
Williams, P.J., Strauss, C.R., Wilson, B. and Massy-Westropp, R.A., 1982. Use of C18 reversed-phase liquid chromatography for the isolation of monoterpene glycosides and nor-isoprenoid precursors from grape juice and wines. J. Chromatogr. A. 235, 2, 471-480. [ Links ]
Wilson, K., 2001. Preparation of Genomic DNA from Bacteria. Curr. Protoc. Mol. Biol. Chapter 2, Unit 2.4. [ Links ]
Yeh, Y.F., Chang, S.C., Kuo, H.W., Tong, C.G., Yu, S.M. and Ho, T.H., 2013. A metagenomic approach for the identification and cloning of an endoglucanase from rice straw compost. Gene 519, 2, 360-366. [ Links ]
Submitted for publication: June 2021
Accepted for publication: October 2021
† Present address: Department of Genetics, Faculty of Natural and Agricultural Sciences, University of the Free State, Bloemfontein, South Africa
* Corresponding author: setati@sun.ac.za
Acknowledgements: This work was funded by the Wine Industry Network for Expertise and Technology (Winetech) grant number SU-IWBT-MG-11/01 and the National Research Foundation-Technology and Human Resources for Industry Programme (NRF-THRIP) grant TP2011060600013. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the funding agencies
















