Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Enology and Viticulture
On-line version ISSN 2224-7904
Print version ISSN 0253-939X
S. Afr. J. Enol. Vitic. vol.41 n.2 Stellenbosch 2020
http://dx.doi.org/10.21548/41-2-4066
ARTICLES
Complete Genome Sequencing of Lactobacillus plantarum UNQLp 11 Isolated from a Patagonian Pinot Noir Wine
N.G. IglesiasII; N.S. BrizuelaI; E.E. TymczyszynI; A. HollmannIII; D. Valdés La HensI; L. SemorileI; B.M Bravo-FerradaI, *
ILaboratorio de Microbiología Molecular, Instituto de Microbiología Básica y Aplicada (IMBA), Departamento de Ciencia y Tecnología, Universidad Nacional de Quilmes, Bernal, Argentina
IILaboratorio de Virus Emergentes, Instituto de Microbiología Básica y Aplicada (IMBA), Departamento de Ciencia y Tecnología, Universidad Nacional de Quilmes, Bernal, Argentina
IIILaboratorio de Compuestos Bioactivos, Centro de Investigación en Biofísica Aplicada y Alimentos (CIBAAL), Universidad Nacional de Santiago del Estero - CONICET, 4206 Santiago del Estero, Argentina
ABSTRACT
Lactobacillus plantarum UNQLp 11 strain was isolated from a Patagonian Pinot noir wine at the oldest commercial winery (110 years old) in General Roca, North Patagonia, Argentina, and has demonstrated its ability to survive during winemaking processes and successfully carry out malolactic fermentation. This work aimed to obtain the whole assembled genome of the UNQLp 11 strain, analysing its architecture and the possible functions of the predicted genes from the oenological properties of this strain. The genome size is 3 534 932 bp, with a mean GC content of 44.2%, 3 412 CDS, 80 transposons and 148 tandem repeats. A comparison between the genome size and gene content of 14 Lb. plantarum strains from different origins was performed, and UNQLp 11 exhibited the largest size. The in silico genome-wide analysis allowed us to confirm the existence of genes encoding enzymes involved in the synthesis of several metabolites of oenological interest, in addition to bacteriocins and exopolysaccharides. Furthermore, it is possible to speculate on this strain's adaptation to different environments, as it is able to use diverse substrates for its growth. All these features suggest the potential of UNQLp 11 to be a good starter culture for malolactic fermentation.
Keywords: Lactobacillus plantarum UNQLp 11, complete genome sequence, native Patagonian wine strain, in silico analysis
INTRODUCTION
Lactobacillus plantarum (recently reclassified in it genus as Lactiplantibacillus plantarum, Zheng et al., 2020) is a species belonging to the lactic acid bacteria (LAB) group, which is found in different ecological niches such as vegetables, meat, fish and dairy products, as well as in the gastrointestinal, vaginal and urogenital tracts (Siezen et al., 2010). This species is a facultative heterofermentative LAB that can utilise a broad range of fermentable carbon sources, hence their application in the production of a variety of foods and beverages, as well as to obtain vitamins, bacteriocins, probiotics, antifungals and potential anticaries agents (Evanovich et al., 2019). Genome sequencing and comparative genomics of different Lb. plantarum strains have revealed high genomic diversity, versatility and flexibility, which ensure its success in various niches and applications (Siezen & Van Hylckama Vlieg, 2011; Jiang & Yang, 2018; Evanovich et al., 2019). This versatility and metabolic capacities make this LAB species highly relevant for industrial applications, and many strains are marketed as starter cultures for fermented foods and beverages (Molin, 2001; Weinberg et al, 2004; Luxananil et al, 2009).
In general, LAB have reduced genomes, but Lb. plantarum presents a larger genome with numerous genes that were acquired by horizontal gene transfer (HGT), mainly via mobile elements (Hubert & Kammerer, 1994; Kleerebezem et al., 2003). The habitat diversity of Lb. plantarum might be related to abundant gene functions. resulting in increased genome size (Bringel et al., 2001; Kant et al., 2011). Genomic analysis has facilitated a more comprehensive characterisation of the genetic characteristics of some bacterial strains (Lasek et al., 2017). In the past decade, several LAB strains were subjected to genome sequencing to further assess their physiological functions and environmental adaptation mechanisms. To date, more than 50 complete genome sequences of Lb. plantarum strains are available in the NCBI GenBank database (Yao et al., 2020). Most of these complete genomes correspond to strains with probiotic properties (Kleerebezem et al., 2003; Zhang et al., 2010; Siezen et al., 2010; Liang et al.,, 2019). In contrast, only a few genomes of oenological strains have been published (Lamontanara et al., 2015; Zhao et al., 2016) or are available from the NCBI GenBank database (CP017363.1).
Oenococcus oeni is the main LAB species responsible for the malolactic fermentation (MLF) of wine, which normally follows the alcoholic fermentation (AF) produced by yeasts (Davis et al., 1985). Also, it is probably the LAB species best adapted to overcome the harsh environmental conditions of wine, and therefore represents the majority of commercial MLF starter cultures. Due to global warming, the harvesting of grapes of higher maturity has resulted in higher pH of the musts. Under these high-pH conditions, Lb. plantarum bacteria have shown especially interesting results for their capacity to induce MLF and for minimising the risk of acetic acid production (Krieger-Weber et al., 2020). Some Lb. plantarum strains can also tolerate the high alcohol and SO2 levels normally encountered in wine and are responsible for MLF in various wine regions and cellars (Guzzon et al., 2009; González-Arenzana et al., 2012; Valdés la Hens et al., 2015). Wine pH has been increasing gradually over the past several years, and red wines with a pH of over 3.5 to 3.6 are occurring more frequently (Martínez de Toda & Balda, 2014). At these pH levels, it is possible to observe a very fast growth in various indigenous microorganisms, some of which are spoilage bacteria that can cause a loss of wine quality (Krieger-Weber et al., 2020). Among these species, Lb. plantarum strains have shown the most interesting results for their capacity to induce MLF under conditions of high pH, their facultative heterofermentative properties that avoid acetic acid production from hexose sugars, and their more complex enzymatic profile and different metabolism compared to O. oeni, which could play an important role in the modification of wine aromas (Du Toit et al., 2011; Bravo-Ferrada et al., 2013; Berbegal et al., 2016; Iorizzo et al., 2016; Krieger-Weber et al., 2020).
Valdés la Hens et al. (2015) showed that Merlot and Pinot noir wines from the oldest cellar (more than 100 years old) in the North Patagonian region of Argentina, at different stages of spontaneous MLF, contained both LAB species, namely O. oeni and Lb. plantarum. In particular, the UNQLp 11 strain of Lb. plantarum was isolated from a Patagonian Pinot noir wine (vintage 2012), and has demonstrated its ability to survive AF and successfully carry out MLF (Bravo-Ferrada et al., 2013, 2014; Brizuela et al., 2017, 2018a, 2018b, 2018c, 2019), suggesting its potential to be employed as a starter culture. In addition, this strain can be grown in a low-cost medium and then used as a starter culture without any previous acclimation treatment (Cerdeira et al., 2019).
The aim of this work was to obtain the whole assembled genome of the UNQLp 11 Lb. plantarum strain so as to analyse its architecture and the possible functions of the predicted genes. The genome sequence of this native Patagonian strain will provide us with useful information about the metabolic capabilities required to successfully carry out the MLF process and its ability to adapt to the stressful wine environment. Furthermore, a comparison with genomes of other Lb. plantarum strains, obtained from different sources, will reveal if UNQLp 11 has undergone specific genetic adaptations that allowed it to be a member of the native bacterial microbiome associated with wine grapes of the Patagonian Pinot noir variety, and to survive during the winemaking process, which would exhibit remarkable performance. The control, improvement and innovation of the MLF process demands detailed knowledge of the possible responses of the bacterium during fermentation and processing conditions.
MATERIALS AND METHODS
Strain information
The UNQLp 11 Lb. plantarum strain was obtained from a Patagonian Pinot noir wine (vintage 2012), in which AF and MLF were spontaneous. This wine was produced in a commercial cellar located in General Roca, Río Negro Province, Argentina. UNQLp 11 was chosen for its significant technological and oenological characteristics (Brizuela et al., 2017, 2018a, 2018b, 2019).
Bacterial growth and DNA extraction
The UNQLp 11 strain was grown in MRS broth at 28°C for two days. To obtain DNA, 1 mg/ml of lysozyme with 1% sodium dodecyl sulphate was used. Proteins were removed with 0.1 g/ml of proteinase K, followed by phenol-chloroform-isoamyl alcohol (25:24:1) extraction. Sixteen μg of high-quality genomic DNA was used for library preparation and sequencing (Iglesias et al., 2019).
Genome sequencing, assembly and bioinformatics analysis
A whole-genome shotgun library was constructed using a 20-kb SMRTbell version 1.0 template prep kit, followed by single-molecule real-time (SMRT) sequencing conducted on an RS II (Pacific Biosciences) sequencer (Macrogen). A total of 1 268 593 327 reads (383,24-fold coverage and a polymerase read N50 size of 21 044 bp), with an average length of14 480 bp and an estimated accuracy of 85.5%, were used as input for de novo assembly with the Canu package v1.8 (Koren et al., 2017). The Canu output consisted of a single circular contig without gaps. Prediction of the coding sequences was done with Gene MarkS-2. Replication and terminus origins were identified using GC-skew analysis and ORF orientation shift.
For genome annotation, the NCBI Prokaryotic Genome Annotation Pipeline was used (Chaudhari et al., 2016). Protein function prediction and KEGG analysis were done by Blast2GO 5.1.1 (Götz et al., 2008). The presence/ absence of exclusive genes in specific Lb. plantarum strains was evaluated by BPGA (Bacterial Pan Genome Analysis Pipeline) (Chaudhari et al., 2016). BPGA determines the core (conserved), accessory (dispensable) and unique (strain-specific) gene pool of a species. COG analysis was done by WebMGA (BMC Genomics 2011, 12:444) (Altermann et al., 2005).
RESULTS AND DISCUSSION
General genome features of UNQLp 11 Lb. plantarum strain
The complete genome of Lb. plantarum UNQLp 11 strain (Gene Bank Accession Number CP031140) contains a single, circular chromosome of 3 534 932 bp with an average GC content of 44.2%, and no plasmid structures were found. Its main features are shown in Table 1. In silico analysis revealed the presence of 3 412 ORFs, 83% of which showed similarities to classified genes from other organisms, and 17% remained unknown. The UNQLp 11 genome also contains 80 transposons, 148 tandem repeats, five ribosomal RNA operons (rRNA), one clustered regularly interspaced short palindromic repeat (CRISPR) locus, and four ncRNAs. CRISPR loci represent a family of DNA repeats typically composed of short and highly conserved sequences (~30 bp), interspaced by variable sequences, often found adjacent to cas genes (CRISPR associated), which are involved in defence systems against the invasion of foreign genetic material, in particular phages (Barrangou et al., 2007). They were also observed in 40% of the sequenced bacterial genomes, and their presence may increase the genome stability, and therefore the environmental adaptation, of bacteria (Sorek et al., 2008).
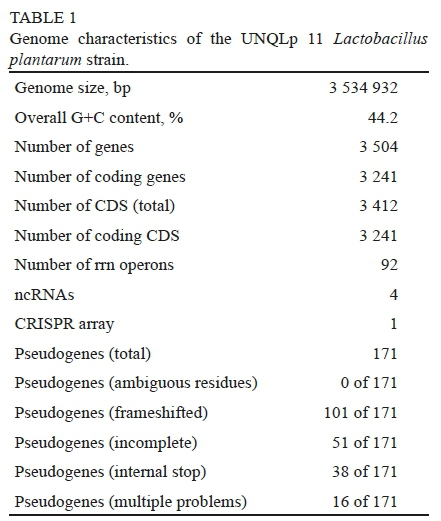
Fig. 1 shows the chromosome atlas of the UNQLp 11 strain. Replication and terminus origins were identified and it was observed that they were located in almost symmetric chromosome positions. Genes encoded in the UNQLp 11 genome are predominantly transcribed in the replication direction, a feature observed in many Gram-positive genomes with low GC content (Kleerebezem et al., 2003).
Fig. 2 shows a comparison between genome size and gene content in 14 Lb. plantarum strains from different origins (plasmid genes present in other strains were not evaluated). UNQLp 11 exhibited the largest size and contains 342 genes more than WCFS1 (control), 405 more than XJ25, 341 more than Lp90, 740 more than Zhang-LL, and 282 more than TMW 1.277 (strains of oenological origin). Higher gene count is not necessarily indicative of transcription followed by a translation into functional proteins.
The UNQLp 11 genome contains 79.3% of protein-encoding genes of the COG class that are involved in the major metabolic pathways and could be assigned to 25 functional categories. Annotation of the UNQLp 11 genome sequence revealed the presence of genes mainly corresponded to the categories transcription, replication, recombination and repair, cell wall/membrane/envelope biogenesis, carbohydrate transport and metabolism. The remaining genes were catalogued as unknown functional proteins. Fig. 3 shows the relative abundance of each COG category of the oenological strains and the control, namely UNQLp 11, Zhang-LL, Lp90, XJ25 and WCFS1.
Furthermore, comparative genomics (gene content) analysis of some Lb. plantarum strains isolated from different fermented foods is shown as a cladogram (Fig. 4). It is evident that the information contained in the genomes of the strains of the Lb. plantarum species of oenological origin cannot determine a relationship between them. It is currently accepted that properties of potential oenological significance are strain-dependent, and the genomic bases have not yet been elucidated (Spano et al., 2006; Capozzi et al., 2012; Bravo-Ferrada et al., 2013). Further studies on the gene regulation mechanisms of Lb. plantarum species are required.
In order to evaluate the similarity between the genomes of Lb. plantarum strains isolated from fermented beverages, we compared the protein coding genes of UNQLp 11 with WCFS1 (reference strain from human saliva), XJ25 (from red wine), Lp 90 (from red wine), Zhang-LL (from rice wine) and TMW1277 (from palm wine). This analysis revealed that the compared genomes share only 2 115 core genes (data not shown).
The analysis of unique genes revealed that they belonged to the categories transcription, replication, recombination and repair, and transport and metabolism of carbohydrates. These results allowed us to assume that a greater gene number in these mentioned categories could increase the ability of UNQLp 11 to employ different sources of nutrients, and to better regulate the transcription processes. UNQLp 11 contains 325 unique genes (10% of its genome), and the smallest percentage of exclusively absent genes (0.1%). These data show that, among the compared genomes, UNQLp 11 contains the highest number of single proteins coding genes and the lowest number of exclusively absent genes (Fig. 5A). A COG analysis of the 10% of unique genes showed that 25% of them could not be classified in any of the COG categories (Fig. 5B). Furthermore, 13% of the 325 unique genes correspond to phage-related genes.
Among the strain-specific genes found in the UNQLp 11 genome are those coding for ATP-dependent Clp proteases, alpha-glucosidases, ABC transporters, PTS (sugar phosphotransferase system) sugar transporters, and USP (universal stress protein). ATP-dependent Clp proteases are involved in several cellular processes, such as degradation of misfolded proteins, regulation of short-lived proteins and housekeeping removal of dysfunctional proteins (Porankiewicz et al., 1999). They are also implicated in the control of cell growth and in the stress response in low-GC Gram-positive bacteria (Fiocco et al., 2010; Russo et al., 2012). Alpha-glucosidases have been suggested to be involved in the degradation of yeast-derived macromolecules as a nutrient source for cell growth (Guilloux-Benatier et al., 1993). The expression of the USP protein is associated with better stress responses by mechanisms still poorly understood (Tkaczuk et al., 2013). The PTS and ABC transporters play a central role in the uptake of different sugars, both in Lb. plantarum and many other bacteria, and their presence is linked with a better capture of nutrients from the environment. Another strain-specific gene also found in UNQLp 11 was mutS, which is involved in the bacterium's repair systems (Garcia-Gonzalez et al., 2012).
Finally, we performed a comparative genomic analysis of four complete genomes, namely UNQLp11, Lp90, Zhang-LL and XJ25, and the reference strain WCFS1, which showed that genes found only in the oenological strains belong to the category of transport and metabolism of carbohydrates. An interesting finding, in the UNQLp 11 genome, was the identification of 27 genes belonging to the family of regulatory proteins of the xenobiotic-sensitive element (Xre), 17 more than in the reference strain, WCFS1 (data not shown). Xre is a critical regulator when the bacterium is in stressful environments, e.g. related to acid tolerance in Lb. acidophilus (Azcárate-Peril et al., 2004) and to ethanol levels in O. oeni (Olguín et al., 2015).
Genes encoding proteins related to carbohydrate transport and metabolism
The Lb. plantarum species, due to its facultatively heterofermentative properties, is heterofermentative for pentoses and homofermentative for hexoses (Dick, 2006). Wine contains many monosaccharides and disaccharides, with glucose, fructose and arabinose being the main sugars metabolised by this LAB species (Hedberg et al., 2008). Numerous genes related to carbohydrate transport and metabolism were found in the UNQLp 11 genome, including 67 phosphoenol pyruvate sugar-transferase systems (PTS) and five sugar-ABC transporters. Among the PTS systems, six families of PTS permeases were identified, namely PTS glucose-glucoside (Glc), PTS fructose-mannitol (Fru), PTS lactose-N,N0-diacetylchitobiose-b-glucoside (Lac), PTS glucitol (Gut), PTS galactitol (Gat), and PTS mannose-fructose-sorbose (Man). As expected, the genome analysis of UNQLp 11 revealed the presence of genes corresponding to whole phosphoketolase and glycolysis pathways. This genome contains five transduction sites (locus tags = DVH03 15355, DVH03 16150, DVH03 06425, DVH03 07445, DVH03 11680) for the L-lactate dehydrogenase protein. This observation supports the relevance of pyruvate-dissipating capacity in this strain.
During the catabolism of readily fermentable sugars such as glucose, the synthesis of enzymes involved in the catabolism of other sugars is repressed, a global regulatory phenomenon termed carbon catabolite repression (CCR) (Bruckner & Titgemeyer, 2002). The dominant mechanism of global carbon control in low-GC Gram-positive bacteria is one involving HPr, the bifunctional HPr kinase/phosphatase (HPrK), and the catabolite control protein A, CcpA (Henkin et al., 1991). CcpA is a member of the Lacl-GalR family of bacterial regulator proteins and regulates the transcription of genes at a global level by binding to a cis-acting DNA sequence designated a catabolite responsive element (cre) (Hueck & Hillen, 1995; Miwa et al, 2000). Catabolite control by CcpA involves both transcriptional activation and repression, and the CcpA regulon is commonly scattered throughout the entire bacterial genome, which has been confirmed by the comparative whole-genome transcriptome analyses in the Gram-positive species Bacillus subtilis, Lb. lactis, and Lb. plantarum, and their ccpA mutant derivatives (Titgemeyer & Hillen, 2002). Also identified were coding genes for the regulatory proteins HPr, CcpA, and HPrK/P (locus tags = DVH03 11560, DVH03 14365, DVH03 06855).
Genes encoding proteins related to amino acid biosynthesis
LAB generally inhabit protein-rich environments (including milk) and are equipped with protein-degradation machinery to create a selective advantage for growth under these conditions (Kleerebezem et al., 2003). Lb. plantarum has uptake systems (Opp and Dtp) for peptides and, once internalised, these peptides are degraded by a variety of peptidases, which have been studied extensively in lactococci and lactobacilli (Ferain et al., 1996). Despite this elaborate protein-degradation machinery, the UNQLp 11 genome contains the genes for the whole biosynthetic pathways of 13 amino acids, namely alanine, aspartate, glutamine, lysine, arginine, cysteine, isoleucine, leucine, methionine, serine, threonine, valine and glycine. For the remaining seven amino acids (asparagine, glutamate, histidine, phenylalanine, tryptophan, tyrosine and proline), the biosynthetic pathways were incomplete, according to the KEGG analysis of maps (data not shown).
Genes encoding proteins related to flavour-development enzymes
Lb. plantarum has a diverse array of enzymes that could have positive effects on aromatic and sensorial wine properties (Du Toit et al., 2011). Besides the malolactic enzyme itself (locus tags = DVH03_12300, DVH03_12380 in the UNQLp 11 genome), other enzymes involved in flavour development include glycosidases, esterases, phenolic acid decarboxylases and citrate lyases (Matthews et al., 2004; Grimaldi et al., 2005). Some (polygalacturonase, pectin methyl esterase) are involved in colour improvement in red wines, or reduce problems associated with wine filtration, such as tannase.
An in silico analysis was performed to look for genes encoding aminopeptidases, glutamate dehydrogenase and phosphotransacylase, among other enzymes (Table 2). Genes that code for alcohol dehydrogenase (AlcDH) and aldehyde dehydrogenase (AldDH) were identified. These enzymes catalyse the conversion of aldehydes to alcohols (AlcDH) or to carboxylic acids (AldDH), which are involved in the fruit flavour development in red wines (Styger et al., 2011). Likewise, the gene coding PAD (phenolic acid decarboxylase) was also found in the UNQLp 11 genome (Table 2, EC: 4.1.1.102). This enzyme participates in the metabolism of phenolic acids from must and wine (Cavin et al., 1993; Swiegers et al., 2005; Mtshali et al., 2010).
The beta-glucosidase enzyme catalyses the release of different aroma compounds by glycosidic bound cleavage, transforming terpenes, alcohols, fatty acids, etc. from bound to free forms (Grimaldi et al., 2000; Spano et al., 2005) (locus tags = DVH03 01055, DVH03 04430, DVH03 04435, DVH03 15725).
Citric acid is present in the wines in concentrations between 0.1 and 1 g/L. The lactic bacteria of the wine cleave it into a molecule of oxalacetate and another of acetate by way of the citrate lyase enzyme. Oxaloacetate is decarboxylated to pyruvic acid, and diacetyl, which is important from an organoleptic point of view because it gives buttery notes to wine, is formed as an intermediate metabolite in the reductive decarboxylation of pyruvic acid to 2,3- butanediol (Bartowsky & Henschke, 2004; Ribéreau-Gayon et al., 2006).
A typical citrate lyase gene cluster (citC, citE, citF, citX) (locus tags = DVH03 12375, DVH03 12365, DVH03 12360, DVH03 12315) was observed in the UNQLp 11 genome.
The gene coding for tannin acyl hydrolase (tannase) (EC: 3.1.1.20), which catalyses ester bond hydrolysis in hydrolysable tannins, such as tannic acid, releasing glucose and gallic acid (Lekha & Lonsane, 1997), was also found in the UNQLp 11 genome. Microorganisms with tannase activity are currently used in the food industry to remove the tannins responsible for undesirable effects in food processing, such as turbidity in wine or fruit juices (Vaquero et al., 2004).
Genes encoding proteins related to stress responses
UNQLp 11 genome analysis allowed the identification of several genes involved in tolerance mechanisms to survive harsh conditions, as previously described in various works (Jobin et al., 1999a; 1999b; Beltramo et al., 2004; Spano & Massa, 2006; Van Bokhorst-Van de Veen et al., 2011). The genes found include clpX (locus tag = DVH03 07590), clpLP (locus tag = DVH03 14965), and trxA (locus tag = DVH03 14965). In addition, class I heat shock genes (groES, groEL and dnaK/dnaJ operons) (locus tags = DVH03 14475, DVH03 14470, DVH03 08265, DVH03 08260), involved in the disulphide-reducing pathway (trxA and trxB homologs) (locus tags = DVH03_06790, DVH03_14250), as well as genes codifying proteins implicated in the ethyl stress response (HrcA and CtsR) (locus tags = DVH03_08250, DVH03 12730), were also identified.
Bacteria protect themselves from changes in environmental osmolarity by using stretch-activated (or mechano-sensitive) channels that respond to changes in membrane tension when cells expand. In the UNQLp 11 genome, genes encoding two-channel protein families (Voltage-gated ion channel (VIC) (locus tag = DVH03_06830) and large conductance mechano-sensitive channel (MscL) (locus tag = DVH03_11240) were found, as well as some aquaporins (locus tag = DVH03_01360) of the MIP (membrane intrinsic proteins) family, which may function primarily to protect the bacteria against osmotic stress (Biggin & Sansom, 2003). In the work of Bienert et al. (2013), it is observed that the number of genes that encode MIP in Lb. plantarum is the largest of those reported in other BAL species, and is suggested to have potential flexibility for different substrates.
Genes encoding proteins related to exopolysaccharide production
Exopolysaccharides (EPSs) are high-molecular-weight carbohydrate polymers secreted extracellularly by many microorganisms (Zhou et al., 2019). EPS are thought to protect bacterial cells against extreme environmental conditions, such as biotic and/or abiotic stresses, including temperature, light intensity, pH and osmotic stress (Donot et al., 2012). The UNQLp 11 genome contains genes that are associated with surface polysaccharide production (locus tags = DVH03 04750, DVH03 09260, DVH03 15400). It should be noted that EPSs have been reported to be responsible for the high viscosity of musts. Besides, after alcoholic fermentation, EPSs form aggregates that could block the filter system, delay spontaneous clarification by sedimentation and worsen wine filterability (Dimopoulou et al., 2017). The amount and type of EPS depends on fermentation conditions, Lactobacillus strain and the growth stage of the microorganism (De Vuyst & Degeest, 1999; Zannini et al., 2016). EPSs can be degraded by other organisms in a complex microbial consortium of wine (Salazar et al., 2009). For these reasons, EPS production of UNQLp 11 in wine requires more studies.
Genes encoding proteins related to bacteriocins
Bacteriocins are ribosomally synthesised antimicrobial peptides or proteins, are ubiquitous in the microbial world and are produced by both Gram-positive and Gram-negative bacteria, have cationic properties and kill target cells by destabilising the integrity of the inner membrane envelope, causing disruption of the membrane potential and/ or leakage of cellular solutes that eventually leads to cell death (Diep et al., 2009). Different Lb. plantarum strains (NC8, WCFS1, J23 and J51) have been found to harbour mosaic pln loci in their genomes (Diep et al., 2009). In the
UNQLp 11 genome, we have identified a region containing some bacteriocin putative genes (plnJ, plnF and plnE, locus tags = DVH03 15860, DVH03 15815, DVH03 15810 respectively) implicated in the synthesis of the EF and JK plantaricins, and plantamicin A. The pln locus is widespread among Lb. plantarum strains of oenological origin and shows genetic diversity and plasticity (Sáenz et al., 2009). The production of these bacteriocins could generate a competitive advantage over other microorganisms, thus favouring successful implantation in the wine environment.
Genes encoding proteins related to undesirable metabolites
Arginine is the major amino acid present in wine, with grape juice concentrations ranging from a few hundred mg l-1 to approximately 2.5 g l-1 (Liu et al., 1995). One of the major concerns about arginine metabolism by wine LAB is the synthesis of ethyl carbamate (EC) precursors (Arena & De Nadra, 2005). Ethyl carbamate, also referred to urethane, is a known animal carcinogen found in fermented foods and beverages, including wine (Araque et al., 2011). The in silico analysis showed that the Lb. plantarum UNQLp 11 genome does not contain enzymes coding genes involved in EC synthesis.
Also, the synthesis of biogenic amines (BA) by LAB during winemaking processes, should be avoided, because these compounds affect wine quality and acceptability (Lonvaud-Funel, 1999; Moreno-Arribas et al., 2003), and they can also induce a variety of diseases when they are consumed (and absorbed) in high concentrations. Although the worldwide regulation is not uniform, wines containing high amounts of histamine are rejected in certain markets due to recommendations for or suggested limits of this compound (Smit et al., 2008). In our analysis, it was possible to verify that the UNQLp 11 genome does not contain genes encoding the enzymes histidine decarboxylase (EC: 4.1.1.17), putrescine carbamoyltransferase (EC: 2.1.3.6), and tyrosine decarboxylase (EC: 4.1.1.25), which are responsible for the synthesis of the main BA found in wines (histamine, tyrosine, putrescine).
CONCLUSIONS
The analysis of the complete genome of the UNQLp 11 Lb. plantarum oenological strain revealed the presence of genes involved in the transport and catabolism of different sugars, the biosynthesis of 13 amino acids, the synthesis of wine aroma compounds, and the production of exopolysaccharides and bacteriocins. When unique genes were analysed, it was observed that they belonged to the categories transcription, replication, recombination and repair, and carbohydrate transport and metabolism. These findings allow us to speculate on their adaptation to different environments, as they are able to use diverse substrates for their growth.
The genome comparison of UNQLp 11 with fourteen other Lb. plantarum strains from fermented foods and beverages did not allow us to observe any genotypic relationship among the strains isolated from wine, revealing that the compared genomes shared only 2 115 core genes.
Although gene presence does not guarantee its expression during winemaking, the genome sequencing and analysis of the native UNQLp 11 strain allowed us to associate phenotypes expressed by this strain under different winemaking conditions (Bravo-Ferrada et al., 2013, 2014; Brizuela et al, 2017, 2018a, 2018b, 2018c) and will improve our understanding of its performance in future winemaking assays on pilot scale.
LITERATURE CITED
Altermann, E., Russell, W.M., Azcarate-Peril, M.A., Barrangou, R., Buck, B.L., McAuliffe, O., Souther N., Dobson A., Duong T., Callanan M. & Lick, S., 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. 102(11), 39063912. [ Links ]
Araque, I., Reguant, C., Rozès, N. & Bordons, A., 2011. Influence of winelike conditions on arginine utilization by lactic acid bacteria. Int. Microbiol. 14, 225-233. [ Links ]
Arena, M.E. & De Nadra, M.C.M., 2005. Influence of ethanol and low pH on arginine and citrulline metabolism in lactic acid bacteria from wine. Res. Microbiol. 156(8), 858-864. [ Links ]
Azcarate-Peril, M.A., Altermann, E., Hoover-Fitzula, R.L., Cano, R.J. & Klaenhammer, T.R., 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 70(9), 5315-5322. [ Links ]
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D.A. & Horvath, P., 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819), 1709-1712. [ Links ]
Bartowsky, E.J. & Henschke, P.A., 2004. The 'buttery' attribute of wine diacetyl desirability, spoilage and beyond. Int. J. Food Microbiol. 96(3), 235-252. [ Links ]
Beltramo, C., Grandvalet, C., Pierre, F. & Guzzo, J., 2004. Evidence for multiple levels of regulation of Oenococcus oeni clpP-clpL locus expression in response to stress. J. Bacteriol. 186(7), 2200-2205. [ Links ]
Berbegal, C., Pena, N., Russo, P., Grieco, F., Pardo, I., Ferrer, S., Spano G. & Capozzi, V., 2016. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 57, 187-194. [ Links ]
Bienert, G.P., Desguin, B., Chaumont, F. & Hols, P., 2013. Channel-mediated lactic acid transport: A novel function for aquaglyceroporins in bacteria. Biochem. J. 454(3), 559-570. [ Links ]
Biggin, P.C. & Sansom, M.S., 2003. Mechanosensitive channels: Stress relief. Curr. Biol. 13(5), R183-R185. [ Links ]
Bravo-Ferrada, B.M., Hollmann, A., Delfederico, L., Valdés la Hens, D., Caballero, A. & Semorile, L., 2013. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 29(9), 1537-1549. [ Links ]
Bravo-Ferrada, B.M., Tymczyszyn, E.E., Gómez-Zavaglia, A. & Semorile, L., 2014. Effect of acclimation medium on cell viability, membrane integrity and ability to consume malic acid in synthetic wine by oenological Lactobacillus plantarum strains. J. Appl. Microbiol. 116(2), 360-367. [ Links ]
Bringel, F., Quénée, P. & Tailliez, P., 2001. Polyphasic investigation of the diversity within Lactobacillus plantarum related strains revealed two L. plantarum subgroups. Syst. Appl. Microbiol. 24(4), 561-571. [ Links ]
Brizuela, N.S., Bravo-Ferrada, B.M., La Hens, D.V., Hollmann, A., Delfederico, L., Caballero, A., Tymczyszyn, E.E. & Semorile, L., 2017. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT 77, 348-355. [ Links ]
Brizuela, N.S., Bravo-Ferrada, B.M., La Hens, D.V., Hollmann, A., Delfederico, L., Caballero, A., Tymczyszyn E.E. & Semorile, L., 2018a. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT 77, 348-355. [ Links ]
Brizuela, N.S., Bravo-Ferrada, B.M., Pozo-Bayón, M.Á., Semorile, L. & Tymczyszyn, E.E., 2018b. Changes in the volatile profile of Pinot noir wines caused by Patagonian Lactobacillus plantarum and Oenococcus oeni strains. Food Res. Int. 106, 22-28. [ Links ]
Brizuela, N.S., Bravo-Ferrada, B.M., Curilén, Y., Delfederico, L., Caballero, A., Semorile, L.C., Pozo Bayón M.A. & Tymczyszyn, E.E., 2018c. Advantages of using blend cultures of native L. plantarum and O. oeni strains to induce malolactic fermentation of Patagonian Malbec wine. Front. Microbiol. 9, 2109. [ Links ]
Brizuela, N., Tymczyszyn, E.E., Semorile, L.C., La Hens, D.V., Delfederico, L., Hollmann, A. & Bravo-Ferrada, B., 2019. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Elec. J. Biotechnol. 38, 10-18. [ Links ]
Brückner, R. & Titgemeyer, F., 2002. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209(2), 141-148. [ Links ]
Cavin, J.F., Andioc, V., Etievant, P.X. & Divies, C., 1993. Ability of wine lactic acid bacteria to metabolize phenol carboxylic acids. Am. J. Enol. Vitic. 44(1), 76-80. [ Links ]
Capozzi, V., Russo, P., Ladero, V., Fernandez, M., Fiocco, D., Alvarez, M.A., Gieco, F. & Spano, G., 2012. Biogenic amines degradation by Lactobacillus plantarum: Toward a potential application in wine. Front. Microbiol. 3, 122. [ Links ]
Cerdeira, V., Bravo-Ferrada, B.M., Semorile, L. & Tymczyszyn, E.E., 2019. Design of a low-cost culture medium based in whey permeate for biomass production of enological Lactobacillus plantarum strains. Biotech. Progress. 35, e2791. [ Links ]
Chambellon, E., Rijnen, L., Lorquet, F., Gitton, C., Van Hylckama Vlieg, J.E., Wouters, J.A. & Yvon, M., 2009. The D-2-hydroxyacid dehydrogenase incorrectly annotated PanE is the sole reduction system for branched-chain 2-keto acids in Lactococcus lactis. J. Bacteriol. 191(3), 873-881. [ Links ]
Chaudhari, N.M., Gupta, V.K. & Dutta, C., 2016. BPGA-an ultra-fast pangenome analysis pipeline. Sci Rep. 6(1), 1-10. [ Links ]
Davis, C.R., Wibowo, D., Eschenbruch, R., Lee, T.H. & Fleet, G.H., 1985. Practical implications of malolactic fermentation: A review. Am. J. Enol. Viticulture 36(4), 290-301. [ Links ]
De Vuyst, L. & Degeest, B., 1999. Expolysaccharides from lactic acid bacteria: Technological bottlenecks and practical solutions. Macromol. Symp. 140, 31-41. [ Links ]
Dick, B., 2011. Action research literature 2008-2010: Themes and trends. Act. Res. 9(2), 122-143. [ Links ]
Diep, D.B., Straume, D., Kjos, M., Torres, C. & Nes, I.F., 2009. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 30(8), 1562-1574. [ Links ]
Dimopoulou, M., Claisse, O., Dutilh, L., Miot-Sertier, C., Ballestra, P., Lucas, P.M. & Dols-Lafargue, M., 2017. Molecular cloning, expression and characterization of Oenococcus oeni priming glycosyltransferases. Mol. Biotechnol. 59(8), 323-333. [ Links ]
Donot, F., Fontana, A., Baccou, J.C. & Schorr-Galindo, S., 2012. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 87(2), 951-962. [ Links ]
Du Toit, M., Engelbrecht, L., Lerm, E. & Krieger-Weber, S., 2011. Lactobacillus: The next generation of malolactic fermentation starter cultures - An overview. Food Bioproc. Tech. 4(6), 876-906. [ Links ]
Evanovich, E., De Souza Mendon9a Mattos, P.J. & Guerreiro, J.F., 2019. Comparative genomic analysis of Lactobacillus plantarum: An overview. Int. J. Genomics 2019(2), 1-11. [ Links ]
Ferain, T., Schanck, A.N. & Delcour, J., 1996. 13C nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J. Bacteriol. 178(24), 7311-7315. [ Links ]
Fiocco, D., Capozzi, V., Collins, M., Gallone, A., Hols, P., Guzzo, J., Weidmann, S., Rieu, A., Msadek, T. & Spano, G., 2010. Characterization of the CtsR stress response regulon in Lactobacillus plantarum. J. Bacteriol. 192(3), 896-900. [ Links ]
Garcia-Gonzalez, A., Rivera-Rivera, R. & Massey, S.E., 2012. The presence of the DNA repair genes mutM, mutY, mutL, and mutS is related to proteome size in bacterial genomes. Front. Genet. 3, 3. [ Links ]
González-Arenzana, L., López, R., Santamaría, P., Tenorio, C. & López-Alfaro, I., 2012. Dynamics of indigenous lactic acid bacteria populations in wine fermentations from La Rioja (Spain) during three vintages. Microb. Ecol. 63(1), 12-19. [ Links ]
Götz, S., García-Gómez, J.M., Terol, J., Williams, T.D., Nagaraj, S.H., Nueda, M.J., Robles, M., Talón, M., Dopazo, J. & Conesa, A., 2008. High- throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10), 3420-3435. [ Links ]
Grant, J.R. & Stothard, P., 2008. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 36(2), 181-184. [ Links ]
Grimaldi, A., Bartowsky, E. & Jiranek, V., 2005. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 99(5), 1061-1069. [ Links ]
Grimaldi, A., McLean, H. & Jiranek, V., 2000. Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacterium, Oenococcus oeni. Am. J. Enol. Viticulture 51(4), 362-369. [ Links ]
Guilloux-Benatier, M., Son, H.S., Bouier, S. & Feuillat, M., 1993. Activités enzymatiques: glycosidases et peptidase chez. Vitis 32, 51-57 [ Links ]
Guzzon, R., Poznanski, E., Conterno, L., Vagnoli, P., Krieger-Weber, S. & Cavazza, A., 2009. Selection of a new highly resistant strain for malolactic fermentation under difficult conditions. S. Afr. J. Enol. Vitic. 30(2), 133141. [ Links ]
Hedberg, M., Hasslöf, P., Sjöström, I., Twetman, S. & Stecksén-Blicks, C., 2008. Sugar fermentation in probiotic bacteria - An in vitro study. Oral Microbiol. Immunol. 23(6), 482-485. [ Links ]
Henkin, T.M., Grundy, F.J., Nicholson, W.L. & Chambliss, G.H., 1991. Catabolite repression of α amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5(3), 575-584. [ Links ]
Hubert, J.C. & Kammerer, B., 1994. Determination of chromosome size and number of rrn loci in Lactobacillus plantarum by pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 120(1-2), 51-56. [ Links ]
Hueck, C.J. & Hillen, W., 1995. Catabolite repression in Bacillus subtilis: A global regulatory mechanism for the Gram-positive bacteria? Mol. Microbiol. 15(3), 395-401. [ Links ]
Iglesias, N.G., Valdés la Hens, D., Olguín, N.T., Bravo-Ferrada, B.M., Brizuela, N.S., Tymczyszyn, E.E., Caballero, A. & Semorile, L., 2018. Genome sequence of Oenococcus oeni UNQOe19, the first fully assembled genome sequence of a Patagonian psychrotrophic oenological strain. Microbiol. Resour. Announc. 7(5),.e00889-18. [ Links ]
Iorizzo, M., Testa, B., Lombardi, S.J., García-Ruiz, A., Munoz-González, C., Bartolomé, B. & Moreno-Arribas, M.V., 2016. Selection and technological potential of Lactobacillus plantarum bacteria suitable for wine malolactic fermentation and grape aroma release. LWT 73, 557-566. [ Links ]
Jiang, Y. & Yang, Z., 2018. A functional and genetic overview of exopolysaccharides produced by Lactobacillus plantarum. J. Funct. Foods 47, 229-240. [ Links ]
Jobin, M.P., Garmyn, D., Diviès, C. & Guzzo, J., 1999a. The Oenococcus oeni clpX homologue is a heat shock gene preferentially expressed in exponential growth phase. J. Bacteriol. 181(21), 6634-6641. [ Links ]
Jobin, M.P., Garmyn, D., Diviès, C. & Guzzo, J., 1999b. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiol. 145(5), 1245-1251. [ Links ]
Kant, R., Blom, J., Palva, A., Siezen, R.J. & De Vos, W.M., 2011. Comparative genomics of Lactobacillus. Microb. Biotechnol. 4(3), 323-332. [ Links ]
Kleerebezem, M., Boekhorst, J., Van Kranenburg, R., Molenaar, D., Kuipers, O.P., Leer, R., Tarchini, R., Peters, S.A., Sandbrink, H.M., Fiers, M.W.E.J., Stiekema, W., Klein Lankhorst, R.M., Bron, P.A., Hoffer, S.A., Nierop Groot, M.N., Kerkhoven, R, De Vries, M., Ursing, B., De Vos, W.M. & Siezen, R.J., 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. 100(4), 1990-1995. [ Links ]
Koren, S., Walenz, B.P., Berlin, K., Miller, J.R., Bergman, N.H. & Phillippy, A.M., 2017. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5), 722-736. [ Links ]
Lamontanara, A., Caggianiello, G., Orrú, L., Capozzi, V, Michelotti, V, Bayjanov, J.R., Renckens, B., Van Hijum, S.A.F.T., Cattivelli, L. & Spano, G., 2015. Draft genome sequence of Lactobacillus plantarum Lp90 isolated from wine. Genome Announc. 3(2), e00097-15 [ Links ]
Lasek, R., Dziewit, L., Ciok, A., Decewicz, P., Romaniuk, K., Jedrys, Z., Wibberg, D., Schlüter, A., Pühler, A. & Bartosik, D., 2017. Genome content, metabolic pathways and biotechnological potential of the psychrophilic Arctic bacterium Psychrobacter sp. DABAL43B, a source and a host of novel Psychrobacter-specific vectors. J. Biotechnol. 263, 64-74. [ Links ]
Lekha, P.K. & Lonsane, B.K., 1997. Production and application of tannin acyl hydrolase: State of the art. Adv. Appl. Microbiol. 44, 216-260. [ Links ]
Liang, Y., Liang, S., Zhang, Y., Deng, Y., He, Y., Chen, Y., Chan Liu, C., Chenli Lin, C. & Yang, Q., 2019. Oral administration of compound probiotics ameliorates HFD-induced gut microbe dysbiosis and chronic metabolic inflammation via the G protein-coupled receptor 43 in non-alcoholic fatty liver disease rats. Probiotics Antimicrob. Proteins 11(1), 175-185. [ Links ]
Liu, M., Nauta, A., Francke, C. & Siezen, R.J., 2008. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl. Environ. Microbiol. 74(15), 4590-4600. [ Links ]
Liu, S., Pritchard, G.G., Hardman, M.J. & Pilone, G.J., 1995. Occurrence of arginine deiminase pathway enzymes in arginine catabolism by wine lactic acid bacteria. Appl. Environ. Microbiol. 61(1), 310-316. [ Links ]
Lonvaud-Funel, A., 1999. Lactic acid bacteria in the quality improvement and depreciation of wine. In: Konings, W. N., Kuipers, O. P. & Huis in 't Veld J.H.J. (eds.). Lactic acid bacteria: Genetics, metabolism and applications. September 1999. Springer, Dordrecht. pp. 317 - 33. [ Links ]
Luxananil, P., Promchai, R., Wanasen, S., Kamdee, S., Thepkasikul, P., Plengvidhya, V., Visessanguan, W. & Valyasevi, R., 2009. Monitoring Lactobacillus plantarum BCC 9546 starter culture during fermentation of Nham, a traditional Thai pork sausage. Int. J. Food Microbiol. 129(3), 312-315. [ Links ]
Martínez de Toda, F. & Balda, P., 2014. Reducing the pH of wine by increasing grape sunlight exposure: A method to mitigate the effects of climate warming. Vitis 53(1), 17-20. [ Links ]
Matthews, A., Grimaldi, A., Walker, M., Bartowsky, E., Grbin, P. & Jiranek, V., 2004. Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 70(10), 5715-5731. [ Links ]
Miwa, Y., Nakata, A., Ogiwara, A., Yamamoto, M. & Fujita, Y., 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28(5), 1206-1210. [ Links ]
Molin, G., 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am. J. Clin. Nutr. 73(2), 380s-385s. [ Links ]
Moreno-Arribas, M.V., Polo, M.C., Jorganes, F. & Munoz, R., 2003. Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 84(1), 117-123. [ Links ]
Mtshali, P.S., Divol, B., Van Rensburg, P. & Du Toit, M., 2010. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 108(4), 1389-1397. [ Links ]
Olguín, N., Champomier-Verges, M., Anglade, P., Baraige, F., Cordero-Otero, R., Bordons, A. & Reguant, C., 2015. Transcriptomic and proteomic analysis of Oenococcus oeni PSU-1 response to ethanol shock. Food Microbiol. 51, 87-95. [ Links ]
Porankiewicz, J., Wang, J. & Clarke, A.K., 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32(3), 449-458. [ Links ]
Ribéreau-Gayon, P., Glories, Y., Maujean, A. & Dubourdieu, D., 2006. The microbiology of wine and vinifications. In: Handbook of enology, vol. 1. John Wiley & Sons, West Sussex. [ Links ]
Rodriguez, H., De las Rivas, B., Gómez-Cordovés, C. & Munoz, R., 2008. Characterization of tannase activity in cell-free extracts of Lactobacillus plantarum CECT 748T. Int. J. Food Microbiol. 121(1), 92-98. [ Links ]
Russo, P., De la Luz Mohedano, M., Capozzi, V., De Palencia, P.F., López, P., Spano, G. & Fiocco, D., 2012. Comparative proteomic analysis of Lactobacillus plantarum WCFS1 and ActsR mutant strains under physiological and heat stress conditions. Int. J. Mol. Sci. 13(9), 1068010696. [ Links ]
Salazar, N., Prieto, A., Leal, J.A., Mayo, B., Bada-Gancedo, J.C., de Los Reyes-Gavilán, C.G. & Ruas-Madiedo, P., 2009. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 92(9), 4158-4168. [ Links ]
Sáenz, Y., Rojo-Bezares, B., Navarro, L., Díez, L., Somalo, S., Zarazaga, M. & Torres, C., 2009. Genetic diversity of the pln locus among oenological Lactobacillus plantarum strains. Int. J. Food Microbiol. 134(3), 176-183. [ Links ]
Siezen, R.J. & Van Hylckama Vlieg, J.E., 2011. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 10(1), 1-10. [ Links ]
Siezen, R.J., Tzeneva, V.A., Castioni, A., Wels, M., Phan, H.T., Rademaker, J.L., Starrenburg, M.J.C., Kleerebezem, M., Molenaar, D. & Van Hylckama Vlieg, J.E., 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 12(3), 758-773. [ Links ]
Sorek, R., Kunin, V. & Hugenholtz, P., 2008. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nature Rev. Microbiol. 6(3), 181-6. [ Links ]
Spano, G., Rinaldi, A., Ugliano, M., Moio, L., Beneduce, L. & Massa, S., 2005. A β-glucosidase gene isolated from wine Lactobacillus plantarum is regulated by abiotic stresses. J. Appl. Microbiol. 98(4), 855-861. [ Links ]
Spano, G. & Massa, S., 2006. Environmental stress response in wine lactic acid bacteria: Beyond Bacillus subtilis. Crit. Rev. Microbiol. 32(2), 77-86. [ Links ]
Styger, G., Prior, B. & Bauer, F.F., 2011. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 38(9), 1145-1159. [ Links ]
Swiegers, J.H., Bartowsky, E.J., Henschke, P.A. & Pretorius, I., 2005. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11(2), 139-173. [ Links ]
Smit, A.Y., Du Toit, W.J. & Du Toit, M., 2008. Biogenic amines in wine: Understanding the headache. S. Afr. J. Enol. Vitic. 29, 109-127. [ Links ]
Titgemeyer, F. & Hillen, W., 2002. Global control of sugar metabolism: A gram-positive solution. In: Lactic acid bacteria: Genetics, metabolism and applications. Springer, Dordrecht. pp. 59 - 71. [ Links ]
Tkaczuk, K.L., Shumilin, I., Chruszcz, M., Evdokimova, E., Savchenko, A. & Minor, W., 2013. Structural and functional insight into the universal stress protein family. Evol. Appl. 6(3), 434-449. [ Links ]
Valdés la Hens, D., Bravo-Ferrada, B.M., Delfederico, L., Caballero, A.C. & Semorile, L.C., 2015. Prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation in Patagonian red wines revealed by polymerase chain reaction-denaturing gradient gel electrophoresis with two targeted genes. Aust. J. Grape Wine Res. 21(1), 49-56. [ Links ]
Van Bokhorst-Van de Veen, H., Abee, T., Tempelaars, M., Bron, P.A., Kleerebezem, M. & Marco, M.L., 2011. Short- and long-term adaptation to ethanol stress and its cross-protective consequences in Lactobacillus plantarum. Appl. Environ. Microbiol. 77(15), 5247-5256. [ Links ]
Vaquero, I., Marcobal, Á. & Munoz, R., 2004. Tannase activity by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 96(2), 199-204. [ Links ]
Weinberg, Z.G., Muck, R.E., Weimer, P.J., Chen, Y. & Gamburg, M., 2004. Lactic acid bacteria used in inoculants for silage as probiotics for ruminants. Appl. Biochem. Biotechnol. 118(1-3), 1-9. [ Links ]
Yao, W., Yang, L., Shao, Z., Xie, L. & Chen, L., 2020. Identification of salt tolerance-related genes of Lactobacillus plantarum D31 and T9 strains by genomic analysis. Ann. Microbiol. 70(1), 1-14. [ Links ]
Zannini, E., Waters, D.M., Coffey, A. & Arendt, E.K., 2016. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 100(3), 1121-1135. [ Links ]
Zhao, M., Liu, S., He, L. & Tian, Y., 2016. Draft genome sequence of Lactobacillus plantarum XJ25 isolated from Chinese red wine. Genome Announc. 4(6), e01216-16. [ Links ]
Zhang, W., Yu, D., Sun, Z., Wu, R., Chen, X., Chen, W., Meng, H., Hu, S. & Zhang, H., 2010. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J. Bacteriol. 192(19), 5268-5269. [ Links ]
Zheng, J., Wittouck, S., Salvetti, E., Franz, C.M., Harris, H.M., Mattarelli, P., O'Toole, P.W, Pot, B., Vandamme, P., Walter, J., Watanabe, K., Wuyts, S., Felis, G.E, Ganzle, M.G. & Lebeer, S., 2020. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70(4), 2782-2858. [ Links ]
Zhou, Y., Cui, Y. & Qu, X., 2019. Exopolysaccharides of lactic acid bacteria: Structure, bioactivity and associations: A review. Carbohydr. Polym. 207, 317-332. [ Links ]
Submitted for publication: May 2020
Accepted for publication: August 2020
* Corresponding author: E-mail address: bbferrada@unq.edu.ar
Acknowledgements: This work was funded by grants from Universidad Nacional de Quilmes (Programa I+D Microbiología Molecular Básica y Aplicada a Agronomía, Alimentos y Salud 2019-2023), the Comisión Nacional de Investigaciones Científicas y Tecnológicas (CIC-BA, Argentina) - Proyecto PIT-AP-BA N° 173 (2016-2019), the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT-MINCyT, Argentina, PICT 2013-1481, 2014-1395, and PICT 2016-3435). NB has a fellowship from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). NGI, BMBF, EET and AH are members of the Research Career of CONICET. LS and DVH are members of the Research Career of Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CIC-BA, Argentina)














