Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Enology and Viticulture
On-line version ISSN 2224-7904
Print version ISSN 0253-939X
S. Afr. J. Enol. Vitic. vol.39 n.2 Stellenbosch 2018
http://dx.doi.org/10.21548/39-2-2685
ORIGINAL RESEARCH ARTICLES
Confirmation of the Effectiveness and Genetic Positions of Disease Resistance Loci in 'Kishmish Vatkana' (Renl) and 'Villard Blanc' (Ren3 and Rpv3)
R. VeikondisI; P. BurgerII; A. VermeulenII; C.J. van HeerdenI; R. PrinsI, III, *
IDepartment of Genetics, Stellenbosch University, Private Bag X1, Matieland, 7602, South Africa
IIARC Infruitec-Nietvoorbij, Private Bag X5026, Stellenbosch 7599, South Africa
IIICenGen (Pty) Ltd, 78 Fairbairn Street, Worcester, 6850, South Africa
ABSTRACT
This study aimed to validate the effectiveness and to genetically characterise the fungal disease resistance genes of 'Kishmish Vatkana' and 'Villard Blanc' in South Africa using microsatellite (SSR) markers and a Quantitative Trait Loci (QTL) approach. An Ft'Sunred Seedless' x 'Kishmish Vatkana' cross was used to generate a partial linkage map for chromosome 13 known to harbour the Renl powdery mildew locus of 'Kishmish Vatkana'. The effectiveness of this locus was validated, explaining between 44.8% and 57.7% of the observed phenotypic variance. An Ft'Villard Blanc' x 'G1-6604' cross was used to generate partial linkage maps for chromosomes 15 and 18, reported to harbour fungal resistance genes of 'Villard Blanc'. The powdery mildew QTL (Ren3) was validated on chromosome 15 of 'Villard Blanc', which explained between 18.9% and 23.9% of the phenotypic variance observed. The downy mildew resistance QTL on chromosome 18 (Rpv3) of 'Villard Blanc' was also confirmed, and it explained between 19.1% and 21.2% of the phenotypic variance observed. This molecular information and individual sources of resistance have already been implemented in the marker-assisted selection (MAS) and gene pyramiding efforts of the table grape breeding program of the Agricultural Research Council (ARC) Infruitec-Nietvoorbij.
Key words: Genetic mapping, QTL, downy and powdery mildew
INTRODUCTION
Grapevine is a very important agricultural plant. It is one of the most widely grown fruit crops in the world and is used for wine making, for fresh consumption, as well as drying. Market demands and producers' requirements for specific traits (e.g. berry quality, taste, yield, shelf life, disease resistance and seedlessness) place constant pressure on breeders to improve plant and fruit traits, which is achieved in conventional breeding programs through progeny selection in inter- or intraspecies crosses (Eibach et al., 2007; Troggio et al., 2008).
The introduction of the insect pest phylloxera and fungal pathogens like downy and powdery mildews from North America had a devastating effect on the vineyards, since the native European grapevines (Vitis vinifera L.) had no inherent resistance. The first documentation of powdery mildew present in Europe was in 1845, while downy mildew was introduced in the late 1870s. In South Africa, powdery and downy mildew was first reported in 1880 and 1907, respectively (Halleen & Holz, 2001; Koopman et al., 2007). This occurrence led to the exploration of the various Vitis species to find resistances that could be introduced into the V. vinifera varieties through the development of introgression lines (Alleweldt et al., 1990; Hoffmann et al., 2008; Gessler et al., 2011). As an alternative approach, various research programs have started to investigate the generation of genetically modified (GM) grapevines, but no commercial cultivars have been released due to strict legislation and strong market resistance to GM plants (Pazzi, 2008; Gray et al., 2014). At present, resistant varieties obtained through conventional breeding are the only acceptable method to produce resistant varieties.
Downy mildew caused by Plasmopara viticola, and powdery mildew caused by Erysiphe necator are two of the bigger threats to growers, as they can lead to major crop and even plant loss, if very severe. Downy mildew infections do not occur every growth season, but it can have devastating effects, depending on the severity of the infection and the phenological stage, when it occurs (Giuntoli & Orlandini, 2000; Fourie, 2003). Powdery mildew in particular is one of the most serious threats, since it is not completely dependent on specific temperature and humidity levels for infection, and it occurs annually in most vineyards (Halleen, 2003; Gadoury et al., 2012).
Using conventional breeding for disease resistance is traditionally a very long and costly process, because of the relative long generation cycle of grapevines. The introduction of molecular markers has made it possible to identify genes or genomic regions related to specific traits, such as disease resistance, much earlier in the breeding cycle. In an attempt to achieve durable resistance, the general hypothesis is that the combination of more than one resistance locus in a single cultivar, which is called gene pyramiding, will ensure that pathogens do not easily overcome the respective resistance genes/QTL (Eibach et al., 2007; Iwata et al., 2016). Extensive research has been done in order to identify the resistance genes or QTL in known resistant varieties and to identify molecular markers linked to these loci, for MAS to improve selection efficiency (Bellin et al., 2009; Barba et al., 2014; Van Heerden et al., 2014).
'Kishmish Vatkana' is a cultivated grapevine documented from Central Asia since the 1920s. It has very good resistance to powdery mildew that has been linked to a single dominant resistance locus to Erysiphe necator (Renl), located on chromosome 13 in a 7.4 centimorgan (cM) interval (Hoffmann et al., 2008; Coleman et al., 2009; Kozma et al., 2009; Riaz et al., 2013). 'Villard Blanc' is an interspecific cross of 'Seibel 6468' and 'Subereux', created by the breeder Bertille Seyve-Villard and is closely related to 'Regent' (Akkurt et al., 2007). On the distal part of chromosome 18, a QTL (Rpv3) is located that is associated with downy mildew resistance (Zyprian et al., 2016). This locus was initially identified in 'Regent' (Fisher et al., 2004; Welter et al., 2007) and named when detected in 'Bianca' (Bellin et al., 2009), a descendent of 'Villard Blanc'. Two loci associated with powdery mildew resistance (Ren3 and Ren9) are located on chromosome 15 of 'Villard Blanc' and 'Regent' (Fisher et al., 2004; Akkurt et al., 2007; Zyprian et al., 2009, 2016; Zendler et al., 2017). In addition to the Ren3 locus, a minor powdery mildew resistance locus (Ren8) is also located on chromosome 18 of 'Villard Blanc' (Zyprian et al., 2016).
In an effort to identify possible sources of resistance to use in local breeding, this study attempted to: 1) validate the effectiveness of previously described fungal resistance QTL of three Vitis varieties under South African conditions, and 2) validate and characterise their chromosomal positions. Foreign disease resistant grapevine varieties were crossed with table grape varieties that are frequently cultivated in South Africa, or with breeding lines developed in the ARC's programme. The resulting F1 progenies were investigated for their inherited disease resistance by identifying markers that are polymorphic between the parental lines. Linkage maps of all relevant linkage groups (LGs) were developed by screening the mapping population with the polymorphic markers. Following phenotypic characterisation of the mapping populations, QTL analyses were performed to identify the respective genomic regions of importance.
MATERIALS AND METHODS
Plant material
A segregating population consisting of 158 F1 individuals from a 'Sunred Seedless' χ 'Kishmish Vatkana' (SS χ KV) cross and a segregating population consisting of 250 F1 individuals from a 'Villard Blanc' χ 'G1-6604' (VB χ G1) cross were used as mapping populations. 'G1-6604' is a breeding line and not a released cultivar.
Molecular analysis
A CTAB extraction method was used to obtain DNA from young leaves of the various populations. Extracted DNA was evaluated using a NanoDrop ND-1000 Spectrophotometer to confirm that the 260/280 ratios were between 1.8 and 2 for all samples. The DNA was then diluted to 30 ng/µL based on the DNA concentrations obtained. The SSR markers chosen for each objective varied, depending on whether a genome scan covering all or selected linkage groups was required, or whether a specific QTL region was targeted. The SSR markers used to screen the 'Villard Blanc' populations were selected from Van Heerden et al. (2014), whilst the 'Kishmish Vatkana' markers were selected from Hoffmann et al. (2008) and the National Centre for Biotechnology Information (NCBI) Map Viewer webpage (www.ncbi.nlm.nih.gov). A standard set of multiplex PCR conditions (1.8 mM MgCl2, 0.75 U Super-Therm Taq, 5mM dNTP and 0.2 pmol/µL of each primer) was used for all reactions. The PCR products were purified using the Macherey Nagel NucleoFast Post PCR purification kit implemented on a Tecan Evo 150 liquid handler using the vacuum protocol provided with the kit. During the PCR purification step, the PCR product was diluted in 70 μL of water. Two microliters of the purified product was separated on an ABI 3730xl using LIZ 500® internal size standard and a 50 cm capillary. The data was scored using GeneMapper® v3.7 and the allele calls exported for formatting prior to mapping.
Disease evaluation
Powdery mildew phenotypic screens were performed on the 'Kishmish Vatkana' and 'Villard Blanc' populations by evaluating disease responses on whole leaves and whole plants with isolates collected in local vineyards with no information on strain identity. V. vinifera vines that were susceptible to powdery mildew and that had naturally occurring infections were identified in the vineyard. A single lesion was collected, from which a single conidium was microscopically picked to infect a potted susceptible V. vinifera plant kept in a growth chamber. This was then used as natural infection source for additional susceptible plants. For the whole leaf scores, two leaves were collected between the fourth and sixth nodes from the shoot tip of each plant. It was rinsed in a 0.35 g/L sodium hypochlorite solution prior to handling in the laboratory and floated adaxial side up on water in a Petri dish, ensuring that the petiole was in the water. Each leaf was inoculated with a few drops of Erysiphe necator suspension (5 x 105 spores/mL). The suspension was generated by collecting conidia from infected, susceptible plants kept in a growth chamber. A leaf infection site was covered with a mixture of water and Tween®, and then lightly brushed with a small brush to release the spores. The liquid was collected and microscopic verification of presence and concentration of spores was done. The Petri dishes were left uncovered for six hours to allow the drops to dry on the leaves before the lids were replaced. The leaves were left for five to seven days in a laboratory where a natural light cycle and constant temperature (24°C) was maintained. Scoring was done according to the OIV 455 criteria for whole leaves and whole plants (Organisation Internationale de la vigne et du vin (OIV), 2007) as 9, 7, 5, 3 or 1, with 9 = very low, greatly suppressed symptoms or none at all - no mycelium or visible fructification (merely a slight curling of leaf blade), and 1 = unlimited infection, complete or nearly complete attack of the leaves - ample mycelium and fungus fructification. For the whole plant scores performed on plants in the greenhouse, the methodology as described by Van Heerden et al. (2014) was followed. V. vinifera vines that were susceptible to powdery mildew and that had naturally occurring infections were identified in the vineyard. A single conidium was microscopically picked from a single lesion to infect a potted susceptible V. vinifera plant kept in a growth chamber. This was then used as natural infection source for additional susceptible plants. These infected plants were then placed amongst the 'Kishmish Vatkana' and 'Villard Blanc' mapping populations in the greenhouse to allow infection to take place. The different parental lines of the mapping populations were included in the phenotypic analysis. After fourteen days, the plants were scored according to the OIV 455 criteria for whole leaves and whole plants (Organisation Internationale de la vigne et du vin (OIV), 2007) as 9, 7, 5, 3 or 1, with 9 = very low, greatly suppressed symptoms or none at all - no mycelium or visible fructification (merely a slight curling of leaf blade), and 1 = unlimited infection, complete or nearly complete attack of the leaves - ample mycelium and fungus fructification.
The downy mildew screen was performed on the 'Villard Blanc' population using a leaf disc assay (Brown et al., 1999). Two leaves were collected between the fourth and sixth nodes from each plant and taken to the laboratory in numbered paper envelopes. The leaves were rinsed in a bleach solution (0.35 g sodium hypochlorite/L) prior to handling in the laboratory to remove all pathogens. Six leaf discs of one cm diameter were excised from each leaf and floated adaxial side up on water in a Petri dish. The six discs per leaf were divided between two Petri dishes so that duplicates were generated to confirm phenotypic scores. Each leaf disc was inoculated with a 10 μL (1.325 x 105 sporangia/mL) drop of Plasmopara viticola suspension. The suspension was generated by collecting spores from infected, susceptible plants kept in a growth chamber. A leaf infection site was covered with a mixture of water and Tween®, and then lightly brushed with a small brush to release the spores. The liquid was collected and microscopic verification of presence and concentration of spores was done. The Petri dishes were left uncovered for six hours to allow the drops to dry on the leaves, before the lids were put on. The leaves were then left for five to seven days in a laboratory where a natural light cycle and constant temperature (24°C) was maintained. When downy mildew growth showed on the discs, they were scored manually using a microscope to determine the level of development of the pathogen. Scoring was done according to criteria of the "Office International de la Vigne et du Vin" (OIV score) for leaf discs (Organisation Internationale de la Vigne et du Vin (OIV), 2009) as 9, 7, 5, 3 or 1, with 9 = very low, tiny necrotic spots, no sporulation nor mycelium, and 1 = very high, strong sporulation and dense mycelium, sporulation bigger than droplet size. The correlation between the different phenotypic scores were determined using R (R Development Core Team 2011), in order to evaluate the correlation (rho value) and the significance of the correlation (P value), if found.
Linkage maps
Although the markers included in the study represented only two different linkage groups, TMAP (Cartwright et al., 2007) and JoinMap v4 (Van Ooijen, 2006; 2011) were used to confirm marker linkage and order. The TMAP software calculated the grouping and order of markers into linkage groups for chromosomes 15 and 18 of 'Villard Blanc', and determined the phasing of each marker. Maps were generated to indicate the order of markers and the distance between them. In JoinMap, the odds ratio of linkage versus no linkage (LOD score) between markers was calculated to determine possible linkage between markers using the Kosambi mapping function. All individuals originating from self-pollination were removed.
QTL analysis
QTL mapping was carried out by using MapQTL v6 (Van Ooijen, 2009), and each phenotypic score was individually examined. Nonparametric Kruskal-Wallis tests were performed on all markers before Interval mapping (IM) was applied to detect putative QTL. The significant LOD threshold for QTL detection (P = 0.05) for each linkage group was determined by 1,000 permutations. Markers that were close to the potential QTL as detected by the Kruskal-Wallis and IM analyses, were chosen as cofactors and tested using automatic cofactor selection criterion in MapQTL. The markers identified as cofactors were used in multiple QTL model (MQM) mapping to determine a more precise location.
RESULTS
'Kishmish Vatkana'
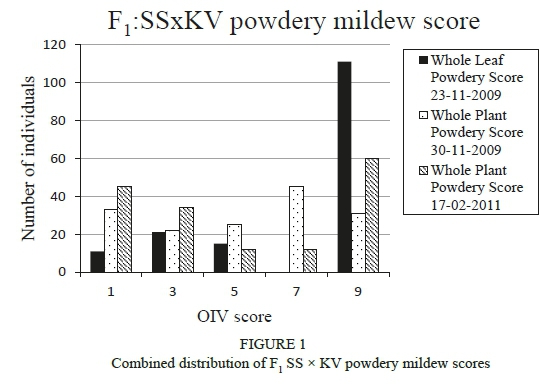
Three powdery phenotypic scores were performed and all the plants that scored 5, 7 and 9 (OIV scores) were considered to be resistant for the purpose of this study. The 23-11-2009 whole leaf score was heavily skewed (79%) towards medium to high levels of resistance (Figure 1). 'Sunred Seedless' (susceptible parent) had a score of 5 and 'Kishmish Vatkana' (resistant parent) had a score of 9. The 30-11-2009 and 1702-2011 whole plant scores had a more even distribution (Figure 1). In both scores, 'Sunred Seedless' was classified as either 1 or 3, while 'Kishmish Vatkana' was classified as 9. The heavily skewed 23-11-2009 whole leaf results were discarded and not used for QTL analysis. Spearman rank and Pearson correlations were performed and both calculations showed rho values of 0.5 and P values of at least 2.0E-12. This confirmed that there was a moderate correlation between the 30-11-2009 and 17-02-2011 scores.
Linkage mapping was performed using eight polymorphic SSR markers and yielded 'Sunred Seedless' chromosome 13 maps of 33.7 cM (TMAP) and 39.4 cM (JoinMap; Figure 2), respectively. The 'Kishmish Vatkana' linkage maps were 37.8 cM (TMAP) and 51.8 cM (JoinMap; Figure 2).
Kruskal-Wallis (single marker regression) analysis revealed that all of the markers on chromosome 13 were significantly associated with the single dominant Renl powdery mildew resistance locus (P<=0.005). The permutation test with 1,000 permutations calculated the significance threshold at a LOD of 2.6 for both phenotypic scores. IM mapping identified a maximum LOD for Renl between markers UDV129 and VVIPP10, with MQM reducing the interval to UDV124 and UDV020 (11.7 cM), which explained up to 57.7% of the phenotypic variance observed (LOD 27.48) (Table 1). The two markers VMC9H4-2 and VMCNG4E10-1 are the most closely linked markers to Renl, followed by UDV-124 and UDV-020 as flanking markers (Figure 3).
'Villard Blanc'
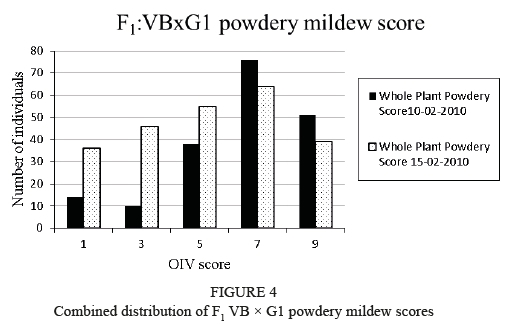
The powdery mildew phenotypic scores were evaluated and all the plants that scored 5, 7 and 9 (OIV scores) were considered to be resistant for the purpose of this study. For the 10-02-2010 whole leaf score, 80% of the F1 VB x G1 population displayed medium to very high levels of resistance (scores of 5 and above), and for the 15-02-2010 whole plant score, 60% displayed levels of medium to very high resistance (Figure 4). The phenotypic scores for the 'Villard Blanc' and 'G1-6604' control plants were very high (OIV 9) and high (OIV 7) for both scores respectively. Spearman rank and Pearson correlations were performed, and both calculations showed rho values of 0.8 and P values of at least 2.2E-16. This confirmed that there was a strong correlation between these two scores with a very high level of significance (P < 0.001).
Linkage mapping was performed with 11 SSR markers of which one marker (VChr15a) was nonpolymorphic in 'Villard Blanc' and five markers (UDV047, VVIQ61, VVIV24, VVIM42b, VMC8G3.2) were nonpolymorphic in 'G1-6604', and were therefore not included in the respective parental maps. The chromosome 15 maps generated for 'Villard Blanc' were 34.3 cM (TMAP) and 35.4 cM (JoinMap), compared to 58.3 cM (TMAP) and 62.6 cM (JoinMap) for 'G1-6604' (Figure 5).
Kruskal-Wallis analysis revealed several markers on chromosome 15 to be associated with resistance to powdery mildew (P<0.005). These associated markers were spanning a 15 cM stretch from UDV116 to VVIP33. A permutation test with 1,000 permutations calculated the significance threshold at a LOD of 2.5 for both phenotypic scores. The position of a QTL linked to powdery mildew resistance (Ren3) of 'Villard Blanc', as calculated with IM, was placed between UDV116 and VVIP33. MQM mapping identified an area of significance located between UDV116 and VVIB63, explaining up to 23.9% of the phenotypic variance observed (Table 1). The QTL interval identified for powdery mildew resistance spans a length of 9.7 cM and the most closely linked markers are UDV116, UDV047 and VVIB63 (Figure 6).
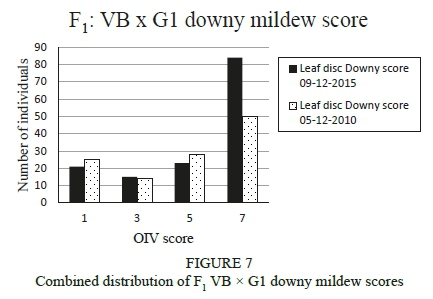
The downy mildew phenotypic scores were evaluated and all the plants that scored between 5 and 9 (OIV score) were regarded as resistant plants. For the 05-122010 score, 66% of the F1 VB x G1 population displayed medium to high levels of resistance, and for the 09-12-2010 score, 71% displayed levels of medium to high resistance (Figure 7). The control plants for these two screens scored a high resistance for 'Villard Blanc' (female parent) and a low to very low resistance for 'G1-6604', the pollen parent. Spearman rank and Pearson correlations were performed, and both calculations showed rho values of 0.8 and P values of at least 2.2E-16. This confirmed that there was a strong correlation between these two scores with a very high level of significance (P < 0.001).
Linkage mapping was performed with 10 SSR markers of which one marker (VVMD17) was nonpolymorphic in 'Villard Blanc' and two markers (VMC8B5, VVIP08) were nonpolymorphic in 'G1-6604'. These nonpolymorphic markers failed to map in the respective parental maps. The chromosome 18 maps generated for 'Villard Blanc' were 85.8 cM (TMAP) and 102.0 cM (JoinMap). The 'G1-6604' linkage maps were 89.1 cM (TMAP) and 101.6 cM (JoinMap) (Figure 8).
Kruskal-Wallis (single marker regression) analysis revealed several markers on chromosome 18 to be associated with resistance to downy mildew (P<0.005). These associated markers were from marker VVIN16-cjvh to VMC7F2, and spans a 7 cM stretch at the distal end of chromosome 18. A permutation test with 1,000 permutations calculated the significance threshold at a LOD of 2.5 for both phenotypic scores. The position of a QTL linked to downy mildew resistance (Rpv3), as calculated with IM, was placed between SSR markers VVMD17 and VMC7F2. MQM mapping identified an area of significance located between markers VMC6F11 and VMC7F2, explaining up to 21.2% of the phenotypic variance observed (Table 1; Figure 9).
DISCUSSION
'Kishmish Vatkana'
The whole leaf phenotypic scoring method for powdery mildew (23-11-2009) produced a very high level of resistance for the whole mapping population. Additionally, no plants with an OIV score of 7 was observed. This could be due to ineffective inoculation events, a laboratory environment that did not promote the growth of the pathogen (temperature and humidity) or low infection pressure due to the time of season or the time between infection and scoring of the population. In addition, the whole leaf scoring process is very labour intensive, which is also problematic when working with bigger populations for fine mapping purposes. For the 30-112009 and 17-02-2011 whole plant scores, all the OIV classes were represented and had a moderate correlation between the scores with a high level of significance. This could be due to the fact that the environment in the nursery tunnels more closely represents the natural environment in terms of factors involved in pathogen spread and growth, and would be the preferred method of screening for powdery mildew resistance in the future.
The linkage maps constructed for 'Sunred Seedless' by using TMAP and JoinMap were fairly similar in length, while there was a significant difference in length for the 'Kishmish Vatkana' map, notably between markers UDV020 and VVIP10. This is most likely due to the difference in the way that the software packages calculate map distances. TMAP is better able to handle incomplete datasets as well as markers that are not completely informative (Cartwright et al., 2007). Since the dataset contained very few missing values, it can only be concluded that the reduced level of informativeness of the markers UDV020 and VVIP10 (both contained shared parental alleles) led to the difference in length observed between the two markers.
MQM successfully validated the presence of the single dominant Renl powdery mildew resistance locus between markers UDV124 and UVD020. The two markers, VMC9H4-2 and VMCNG4E10-1, are the most closely linked markers to the QTL, followed by UDV124 and UDV020 as flanking markers confirming the chromosome location as identified previously (Hoffmann et al., 2008; Coleman et al., 2009; Riaz et al., 2013). Given that the two whole plant scores (30-11-2009 and 17-04-2011) confirmed the effectiveness and genetic position of the powdery mildew resistance Renl locus in the local South African population, it was not deemed necessary to repeat the whole leaf score. Closely linked markers, with the resistance associated alleles were identified, which can be used in the future for MAS breeding purposes and the pyramiding of resistance genes.
'Villard Blanc'
Two powdery mildew phenotypic screens were performed on the F1 VB x G1 population, and for the 10-02-2010 score, 80% of the population displayed medium to very high levels of resistance. For the 15-02-2010 score, 60% displayed levels of medium to very high resistance. The skewness towards resistance could be due to ineffective inoculation events or an environment that did not promote the growth of the pathogen, such as low infection pressure in the greenhouse environment. However, the phenotypic scores for the 'Villard Blanc' and 'G1- 6604' control plants were both very high (OIV 9) and high (OIV 7) for both scores respectively. These high parental scores make it plausible that both were contributing towards resistance, although the major effect was only detected on chromosome 15 of 'Villard Blanc'.
The parental and combined linkage maps constructed for chromosome 15 was similar in length compared to one another, and the combined map was similar to the published map length, whilst retaining the same marker order.
In this study using 250 individuals, we validated the presence of the powdery mildew resistance QTL (Ren3) in 'Villard Blanc' spanning a distance of 8.5 cM between markers UDV116 and UDV047, a region more than 5 cM proximal to VVIP33. This region overlaps the powdery resistance gene/ QTL reported on chromosome 15 by Akkurt et al. (2007) for 'Villard Blanc' in a 'Gf.Ga-47-42' χ 'Villard Blanc' cross. The SCAR markers, associated with the resistance QTL in their study, were developed between markers UDV116 and VVIP33. Zyprian et al. (2016) also mapped a powdery mildew resistance QTL proximal of VVIP33, using the same mapping population as Akkurt et al. (2007). However, the map for chromosome 15 only extends 5.8 cM proximal of VVIP33 and does not include UDV116 or UDV047, which makes direct comparisons problematic. In studies of its close relative, 'Regent', Eibach et al. (2003) and Van Heerden et al. (2014; 206 individuals) validated the position of the powdery mildew resistance locus (Ren3) distal of UDV116. Recently, Zendler et al. (2017) revealed through fine mapping using multiple and larger populations (236 - 1050 individuals), that the Ren3 resistance are actually conferred by two hypersensitive loci in this interval. They proposed that these loci should be designated as Ren3 (interval ScORGF15-02 - ScORA7) and Ren9 (interval CenGen7 - GF15-10). While the loci identified by the various studies above varied slightly in location, the low number of shared markers makes any direct comparison very difficult. Unfortunately, the CenGen6 and CenGen7 markers, previously called VChr15CenGen06 and -07, (Van Heerden et al., 2014), were not included in the initial parental screens and therefore not included in this study.
Two downy mildew phenotypic screens were performed on the F1 VB χ G1 population. The same leaf discs were used for the two screens, as the initial evaluation (05-122010) showed that the infection had not taken place in quite a number of discs. At the time, it was postulated that the laboratory environment was not constant enough, since the temperature and humidity was only controlled during the day. Steps were taken and the air conditioning unit remained on for 24 hours during the infection stage, and a second score was done (09-12-2010). This second score displayed a fairly high percentage of resistance, which could be related to reduced viability of inoculum, ineffective inoculation events or an unfavourable and inconsistent environment where the large number of Petri dishes were kept. However, the control plants for these two screens scored a high resistance for the 'Villard Blanc' parent and a low to very low resistance for the 'G1-6604' parent. The fact that the correlation calculations showed that there was a strong correlation between the two scores performed with a very high level of significance is to be expected, since the scores were taken over a short time span and a second season's phenotypic screen would have been more desirable.
The linkage map for chromosome 18 showed a significant difference in length between the two parental maps, which is most likely due to the difference in the way that the software packages calculate map distances. Seventy percent of the markers on chromosome 18 shared alleles between the parental genotypes and were therefore less informative. TMAP employs a general multipoint-likelihood maximisation method and is more robust in dealing with incomplete datasets and less informative markers (Cartwright et al., 2007). JoinMap (v4.1) employs a multipoint-likelihood maximisation method specific for cross pollinating populations and therefore produces slightly longer maps with marker orders comparable to that generated by TMAP. The combined map for VB χ G1 is very similar to the published map length, since the marker order is the same but the positions of the markers are slightly different, which is expected between different mapping populations.
MQM mapping located a downy mildew QTL between markers VVIN16 and VMC7F2. A QTL for downy mildew resistance (Rpv3) was reported on chromosome 18 in 'Regent' (Eibach et al., 2007; Van Heerden et al., 2014) and 'Villard Blanc' (Zyprian et al., 2016). The QTL position reported by Bellin et al. (2009) was between the SSR markers VMC7F2 and VVIN16, which is the same QTL found in this study. Van Heerden et al. (2014) identified a marker (VChr18c) close to VMC7F2 that can also be considered for future use.
CONCLUSIONS
The effectiveness of the Renl powdery mildew resistance locus of 'Kishmish Vatkana' in South Africa was proven, and closely associated markers for MAS was identified. The presence and position of the genes for powdery mildew and downy mildew resistance were confirmed on chromosomes 15 (most likely Ren3) and 18 (Rpv3) of the 'Villard Blanc' population. The minor effect of Ren8 was not detected. The study laid a foundation for the breeder to eliminate F1 progeny plants, which did not inherit the desired disease resistance, very early in the breeding schedule. By applying these markers in combination with those identified by Van Heerden et al. (2014), the resources spent on carrying unsuitable plants through the breeding cycle could be reduced significantly, and the program could be conducted more efficiently and cost-effectively. The resistance genes and QTL identified in this study can be combined, through conventional breeding techniques, into a single plant to pyramid resistance genes, and thus generate varieties with more durable resistance. This will hopefully result in a reduction of the number of chemical applications that needs to be applied by producers.
LITERATURE CITED
Akkurt, M., Welter, L., Maul, E., Töpfer, R. & Zyprian, E., 2007. Development of SCAR markers linked to powdery mildew (Uncinula necator) resistance in grapevine (Vitis vinifera L and Vitis sp). Mol. Breed. 19, 103-111. [ Links ]
Alleweldt, G., Spiegel-Roy, P. & Reisch, B., 1990. Grapes (Vitis). In: Moore, I.N., Ballington, J.L. (eds). Genetic resources of temperate fruit and nut crops. Acta Hort. 290, 291-327. [ Links ]
Barba, P., Cadle-Davidson, L., Harriman, J., Glaubitz, J.C., Brooks, S., Hyma, K. & Reisch, B., 2014. Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor. Appl. Genet. 127, 73-84. [ Links ]
Brown, M.V., Moore, J.N., Fenn, P. & McNew, R.W., 1999. Comparison of leaf disk, greenhouse and field screening procedures for evaluation of grape seedlings for downy mildew resistance. Hort. Science. 34, 331-333. [ Links ]
Bellin, D., Peressotti, E., Merdinoglu, D., Wiedemann-Merdinoglu, S., Adam-Blondon, A-F., Cipriani, G., Morgante, M., Testolin, R. & Di Gaspero, G., 2009. Resistance to Plasmopara viticola in grapevine 'Bianca' is controlled by a major dominant gene causing necrosis at the infection site. Theor. Appl. Genet. 120, 163-176. [ Links ]
Cartwright, D.A., Troggio, M., Velasco, M. & Gutin, A., 2007. Genetic mapping in the presence of genotyping errors. Genetics. 176, 2521-2527. [ Links ]
Coleman, C., Copetti, D., Cipriani, G., Hoffmann, S., Kozma, P., Kovács, L., Morgante, M., Testolin, R. & Di Gaspero, G., 2009. The powdery resistance gene RENl co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genetics. 10, 89. [ Links ]
Eibach, R. & Töpfer, R., 2003. Success in resistance breeding: 'REGENT' and its steps into the market. Acta Hort. 603, 687-691. [ Links ]
Eibach, R., Zyprian, E., Welter, L.J. & Töpfer, R., 2007. The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis. 46, 120-124. [ Links ]
Fourie, P,. 2003. Downy mildew on grapevine. Vesuvius, Cape Town. [ Links ]
Gadoury, D.M., Cadle-Davidson, L., Wilcox, W.F., Dry, I.B., Seem, R.C. & Milgroom, M.G., 2012. Grapevine powdery mildew (Erysiphe necator): a fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph. Mol. Plant Pathol. 13, 1-16. [ Links ]
Gessler, C., Pertot, I. & Perazzolli, M., 2011. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50, 3-44. [ Links ]
Giuntoli, A. & Orlandini, S., 2000. Effects of downy mildew on photosynthesis of grapevine leaves. Acta Hort. 526, 461-466. [ Links ]
Gray, D.J., Li, Z.T. & Dhekney, S.A., 2014. Precision breeding of grapevine (Vitis vinifera L.) for improved traits. Plant Science. 228, 3-10. [ Links ]
Halleen, F., 2003. Poeieragtige meeldou in wingerd.Vesuvius, Cape Town. [ Links ]
Halleen, F. & Holz, G., 2001. An Overview of the biology, epidemiology and control of Uncinula necator (powdery mildew) on grapevine, with reference to South Africa. S. Afr. J. Enol. Vitic. 22, 111-122. [ Links ]
Hoffmann, S., Di Gaspero, G., Kovács, L., Howard, S., Kiss, E., Galbács, Z., Testolin, R. & Kozma, P., 2008. Resistance to Erysiphe necator in the grapevine 'Kishmish vatkana' is controlled by a single locus through restriction of hyphal growth. Theor. Appl. Genet. 116, 427-238. [ Links ]
Iwata, I., Minamikawa, M.F., Kajiya-Kanegae, H. & Ishimori, M., 2016. Genomics-assisted breeding in fruit trees. Breeding Science. 66, 100-115. [ Links ]
Koopman, T., Linde, C.C., Fourie, P.H. & Mcleod, A., 2007. Population genetic structure of Plasmopara viticola in the Western Cape Province of South Africa. Mol. Plant Pathol. 8, 723-736. [ Links ]
Kozma, P., Kiss, E., Hoffmann, S., Galbács, Z. & Dula, T., 2009. Using the powdery mildew resistant Muscadinia rotundifolia and Vitis vinifera 'Kishmish vatkana' for breeding new cultivars. Acta Hort. 827, 559-564. [ Links ]
Organisation Internationale de la Vigne et du Vin (OIV), 2007. 2nd Edition of the OIV descriptor list for grape varieties and Vitis species. Available at: www.oiv.int/oiv/...%201%20Publications%20OIV/.../5-1-9_Liste_descripteurs_2ed_EN.pdf. [ Links ]
Pazzi, F., 2008. Genetically modified grapevine: state of research, possible risks and future scenario. http://www.fondazionedirittigenetici.org/vitevita/rapporto_en.pdf. [ Links ]
Riaz, S., Boursiquot, J-M., Dangl, G.S., Lacombe, T., Laucou, V., Tenscher, A.C. & Walker, A.M., 2013. Identification of mildew resistance in wild and cultivated Central Asian grape germplasm. BMC Plant Biol. 13, 149. [ Links ]
Troggio, M., Vezzulli, S., Pindo, M., Malacarne, G., Fontana, P., Moreira, F.M., Costantini, L., Grando, M.S., Viola, R. & Velasco, R., 2008. Beyond the genome, opportunities for a modern viticulture: a research overview. Am. J. Enol. Vitic. 59, 117-127. [ Links ]
Van Heerden, C.J., Burger, P., Vermeulen, A. & Prins, R., 2014. Detection of downy and powdery mildew resistance QTL in a 'Regent' χ 'RedGlobe' population. Euphytica. 200, 281-295. [ Links ]
Van Ooijen, J.W., 2006. JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma, B.V. (eds). Wageningen, Netherlands. [ Links ]
Van Ooijen, J.W. 2009. MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma, B.V. (eds). Wageningen, Netherlands. [ Links ]
Van Ooijen, J.W., 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet. Res. Camb. 93, 343-349. [ Links ]
Zendler, D., Schneider, P., Töpfer, R. & Zyprian, E., 2017. Fine mapping of Ren3 reveals two loci mediating hypersensitive response against Erysiphe necator in grapevine. Euphytica. 213, 68. [ Links ]
Zyprian, E., Welter, L.J., Akkurt, M., Töpfer, R., Ebert, S., Salakhutdinov, I., Göktürk-Baydar, N. & Eibach, R., 2009. Genetic analysis of fungal disease resistance in grapevine. Acta Hort. 827, 535-538. [ Links ]
Zyprian, E., Ochßner, I., Schwander, F., Simon, S., Haussmann, L., Bonow-Rex, M., Moreno-Sanz, P., Grando, M.S., Wiedemann-Merdinoglu, S., Merdinoglu, D., Eibach, R. & Töpfer, R., 2016. Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol Genet Genomics. 291, 1573-1594. [ Links ]
Submitted for publication: October 2017
Accepted for publication: August 2018
Acknowledgements: This work is based on the research supported in part by the National Research Foundation (NRF) of South Africa (THRIP grants 70082, 72059 and 75125; NRF Incentive Grant UID 85943). The NRF is also thanked for funding the equipment based at the Central Analytical Facilities of Stellenbosch University (UID 65258). Deciduous Fruit Producers ' Trust (DFPT) and the South African Table Grape Industry (SATI) are thanked for their financial support. Debbie Snyman (CenGen) is thanked for technical assistance. Louise Warnich (Stellenbosch University) is thanked for useful comments and suggestions during the study
* Corresponding author: E-mail address: cengen@cengen.co.za














