Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Industrial Engineering
versão On-line ISSN 2224-7890
versão impressa ISSN 1012-277X
S. Afr. J. Ind. Eng. vol.34 no.3 Pretoria Nov. 2023
http://dx.doi.org/10.7166/34-3-2960
SPECIAL EDITION
A comprehensive overview and evaluation of link prediction techniques
L.M. Brown*; G.S. Nel
Stellenbosch Unit for Operations Research in Engineering, Department of Industrial Engineering, Stellenbosch University, Stellenbosch, South Africa
ABSTRACT
This paper provides a comprehensive overview and evaluation of link prediction techniques. The study includes an analysis of various methods, ranging from simple heuristics to complex embedding-based approaches. The comparative study evaluates the performance of each technique across a range of diverse data sets, and offers unique insights into the strengths and limitations of each approach, as well as their suitability for different types of network structure. For example, the research shows that, while some techniques may perform well on small and sparse networks, they may not be as effective on larger, denser networks. By providing a thorough analysis of various link prediction techniques, this study proffers a valuable resource for researchers seeking to develop more effective algorithms for predicting links in networks. The findings of this study contribute to a deeper understanding of the dynamics and structure of networks.
OPSOMMING
Hierdie artikel bied 'n omvattende oorsig en evaluering van skakelvoorspellingstegnieke. Die studie sluit 'n ontleding van verskeie metodes in, wat wissel van eenvoudige heuristieke tot komplekse inbedding-gebaseerde benaderings. Die vergelykende studie sluit 'n evaluering van die prestasie van elke tegniek oor 'n reeks uiteenlopende datastelle, en bied unieke insigte in die sterk punte en beperkings van elke benadering, sowel as hul geskiktheid vir verskillende tipes netwerkstruktuur. Byvoorbeeld, die bevindinge toon dat, hoewel sommige tegnieke goed kan presteer op klein en yl netwerke, hulle dalk nie so effektief is op groter, digter netwerke nie. Deur 'n deeglike ontleding van verskeie skakelvoorspellingstegnieke te verskaf, bied hierdie studie 'n waardevolle hulpbron vir navorsers wat meer effektiewe algoritmes wil ontwikkel vir die voorspelling van skakels in netwerke. Die bevindinge van hierdie studie dra by tot 'n dieper begrip van die dinamika en struktuur van netwerke.
1. INTRODUCTION
Network science is a powerful tool for understanding complex systems across various inherently interconnected domains, such as social networks, biological systems, and transportation infrastructure, to name a few [1, 2]. A 'network' is formally defined as a system comprising nodes that are connected by links. Nodes represent individual entities or elements, while links represent the connections or relationships between these entities [2]. One of the fundamental tasks in network science is link prediction, which aims to identify potential connections between nodes in a network, based on the current network's connections and other node attributes [3, 4]. Accurate and robust link prediction techniques can prove valuable in extracting insight from networks so as to lend decision support across numerous use cases, ranging from recommending friends in social networks to identifying potential drug targets in biological systems [5, 6].
Over the past few decades, a plethora of link prediction methods has been proposed, encompassing traditional approaches that employ network topology, as well as more sophisticated techniques using machine learning and graph neural networks (GNNs) [6, 7, 8, 9]. Despite the abundance of research in this area, the absence of a comprehensive and unbiased comparative study of a wide range of link prediction methods contributes to inconclusive and inconsistent findings, hindering the identification of the most suitable technique for a specific application [6, 8, 10]. In addition, a large number of evaluation procedures employed in prior studies often disregard crucial considerations that can significantly impact the assessment of link prediction performance, such as the trade-offs between true positive rates and false positive rates, or the impact of the distance between node pairs on algorithmic performance [10]. Furthermore, the efficacy of link prediction techniques can be largely attributed to specific structural properties of the network at hand. Prior studies, however, have often been lacking because of limited insight into this relationship, focusing primarily on heuristic link prediction methods or overlooking certain network structure characteristics [11]. In addition, previous studies typically examined the correlation between a single network characteristic and performance [12, 13]. Recognising the correlative relationship between network structure and prediction performance is key to both selecting the appropriate method for a given application and creating robust prediction techniques.
In this article, the aim is to address these challenges by undertaking a comprehensive and rigorous comparison of a diverse range of link prediction methods. The primary contributions can be summarised as follows:
1. A broad review of state-of-the-art link prediction methods is provided, highlighting their strengths and limitations, while emphasising the lack of a single method that outperforms each of its counterparts in respect of various test problems.
2. A standardised, fair, and effective data set selection and algorithmic evaluation criteria is formulated and applied, focusing on data set diversity and addressing the class imbalance problem that is inherent in link prediction tasks.
3. An extensive comparative study is carried out in respect of the selected techniques on a diverse set of real-world networks, highlighting their strengths and weaknesses and providing practical guidelines for selecting the most appropriate technique according to the specific characteristics of a given network and application domain.
4. A detailed correlation analysis is also performed, investigating the relationship between various network characteristics and the performance of different link prediction techniques. This analysis contributes to a deeper understanding of how the underlying network structure can influence link prediction performance.
5. The importance of fair and effective evaluation is propounded in an effort to advance link prediction research and promote the adoption of these techniques in practical scenarios.
The remainder of this article is organised as follows. A literature review on link prediction methods is presented in Section 2. The methodology - encompassing the selection of link prediction methods, data sets, evaluation criteria, and correlative analysis - is then outlined in Section 3. A detailed analysis and comparison of the link prediction methods is provided in Section 4. The paper concludes in Section 5 with a summary and recommendations for future work.
2. LITERATURE REVIEW
Given a network, denoted by g = (V, £), where V denotes the set of nodes and £ denotes the set of edges (or links), link prediction involves estimating whether a link, denoted by e(u, v), is currently present or is likely to emerge between a pair of nodes u and v, where v, ueV and e(u, v) ¢£. A visual representation of the link prediction problem is presented in Figure 1.
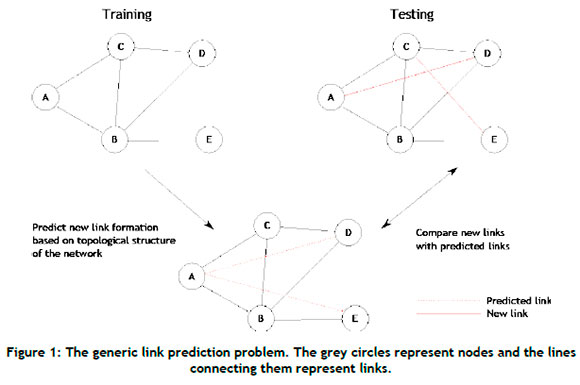
The task of link prediction can generally be described according to two distinct categories [5]:
1. Anticipating future links within a given network, according to which the network's evolving nature underpins the task. This situation relates to tasks such as forecasting future friendships or collaborations, from which valuable insights into the factors that drive network evolution can be gleaned [13].
2. Discerning missing links within an observed network, according to which the network is regarded in its current static state. This approach is largely employed to uncover hidden or lost connections [14, 15].
A wide range of link prediction approaches has been proposed in the literature. These methods can be broadly classified according to three paradigms: heuristic-based, classifier-based, and embedding-based methods [8]. Detailed discussions of each of these categories are presented below.
2.1. Heuristic-based methods
Heuristic-based link prediction methods are governed by the assignment of a so-called similarity score, denoted by S(u,v), to each node pair, which is proportional to the likelihood of a link. These scores are ranked, after which a threshold is employed to determine the specific link predictions. The threshold is often determined empirically or through cross-validation so as to balance the trade-off between precision and recall of the predictions [7]. These methods depend on the network's topological features, and can be classified into three categories: neighbour-based, path-based, and random-walk-based metrics [6].
The common neighbours (CN) metric is a foundational heuristic in the domain of link prediction, predicated on the assumption that two nodes are more likely to form a link if they share a comparatively large number of neighbours [16]. In the context of a network, 'neighbour' refers to a node that is directly connected to another node by a link [2]. For a given pair of nodes u and v, the CN metric is defined as

where Γ(u) and Γ(v) denote the sets of neighbours of u and ν in G = (V,E),respectively. Furthermore, |... | denotes the count of nodes in the neighbourhood.
The Jaccard coefficient (JC) is also based on the principle that shared neighbours between two nodes correspond to a reasonable likelihood of a future link [7, 17]. The JC introduces a normalisation factor to account for the differences in respect of the degrees of the nodes, which provides a more balanced measure. Mathematically, the JC may be defined as

The Salton index (SI) provides a more nuanced measure by incorporating both the intersection and geometric mean of the degrees of the nodes. 'Degree of a node' refers to the number of links connected to that node in a network [2]. The SI accounts for degree variations among nodes, ensuring a more equitable comparison [18]. This can be expressed mathematically as

where ku and kv denote the degrees of nodes u and ν respectively.
The Adamic-Adar (AA) index considers both the quantity of common (i.e., shared) neighbours and the assignment of a weighted importance value. The underlying premise is that shared neighbours that have smaller degrees are more informative; therefore, it assigns a correspondingly smaller weight to shared neighbours having a larger degree [19]. Mathematically, the AA index may be defined as

The preferential attachment (PA) index assumes that nodes having a large degree are more likely to form connections. Accordingly, the probability that a new link involves a particular node is calculated based on its degree, leading to the so-called 'rich get richer' phenomenon [20]. The PA index can be expressed as

The resource allocation (RA) index focuses on resource transfer over the network, which involves the exchange (or sharing) of information, energy, or influence among the nodes in the network [7]. This metric employs the inverse of the degree as the weight [21]. The RA index can be expressed as

Neighbour-based heuristic link prediction methods are commonly associated with advantages such as ease of application and scalability, enabling efficient computation for large-scale networks; the majority of the computational advantages can be attributed to their simple usage of network topology. Their assumption of network homogeneity, however, potentially results in complex structures not being considered satisfactorily, leading to diminished predictive performance [7].
2.2. Learning-based methods
There has been a notable increase in the recent literature in the development of learning-based link prediction methods. These methods integrate heuristic-based link prediction metrics, together with innate properties and other external data [8]. These techniques are typically categorised as follows: classifier-based, probabilistic graph modelling, or matrix factorisation [6].
In the context of classifier-based methods, consider the nodes u, ν e V in the graph G(V,E), and let l(u,v) denote the label for the node pair instance (u, v). The task of learning-based link prediction entails categorising node pairs as instances, which includes assigning a class label to each pair, during which features that describe the characteristics of the pair are used. Therefore, a node pair may be assigned a positive label if a link is present between the nodes. On the other hand, if no such link is present, the pair is labelled as negative. The label of the corresponding data point in the classification model can therefore be defined as

This formulation corresponds to a binary classification task that can be addressed using various supervised learning models [6]. Al-Hasan et al. [3] first proposed a supervised learning approach to link prediction in 2005 - a methodology that may be regarded as a central tenet of the principles of contemporary learning-based methods.
In order to construct an effective link prediction classifier, defining and extracting a set of relevant features from a network is important [3, 7]. The features provided by heuristic-based metrics provide an intuitive means to this end, and are informative in nature. Non-topological features, on the other hand, have the advantage of enhancing the performance of the link prediction problem, but they may not always be easily accessible, thus making collection difficult. Moreover, these features are typically domain-specific, requiring adequate domain knowledge for identification and discovery. While a general link prediction classifier often accounts solely for generic features (such as those related to the node, network, and topological traits), practical link prediction applications should also consider non-topological features to the greatest possible extent [6, 8]. The scope of this study does not permit the inclusion of such features.
Classifier-based link prediction methods are flexible, facilitating the selection of many algorithmic approaches and feature types that can help to capture diverse network structures [22]. Their success, however, can be dependent on feature availability and quality. These approaches can also be computationally demanding, especially for large networks [7]. Consequently, addressing feature challenges and computational complexity is vital for practical application.
2.3. Embedding-based methods
Network embedding-based methods can be equated with dimensionality reduction techniques, according to which high-dimensional nodes in a network are mapped to a lower dimensional representation space while preserving the node characteristics and attributes in a compressed format [8, 23]. Formally, given a network g = (V, £), the aim of network embeddings is to derive a mapping function ƒ: v->rvΕRd, where rv denotes the real-valued vector representation of node v, and d is simply the dimension of the representation [24]. Embedding-based methods can be categorised as follows: matrix factorisation, random walks, and GNNs [8]. In the context of this study, however, focus is placed on random walks and GNNs, given the arguably simpler implementation of the former and the recent popularity of the latter.
In the context of network analysis, random walks involve traversing a network by moving randomly from one node to another, during which network information is collected [25]. DeepWalk is the first random walk embedding-based method to be proposed for link prediction [26]. The approach employs random walks in order to generate node sequences, and employs a skip-gram model [27] from the domain of representation learning (Hamilton, Graph representation learning, 2020). By interpreting node sequences as sentences, latent representations can be approximated, providing a novel perspective for network embedding approaches [7, 26]. Building on the foundation of DeepWalk, Grover et al. [29] proposed Node2Vec. This method introduces a biased random walk strategy that merges breadth-first and depth-first search.
Although random walk-based embeddings can handle large networks efficiently, their utility is contingent on the assumption that node neighbourhoods are indicative of the likely presence of links - an assumption that may not hold in complex networks [6]. The quality of embeddings is also sensitive to the chosen parameter values; these methods can also fail to capture long-range dependencies effectively. It is therefore imperative to evaluate critically the underlying assumptions and limitations when applied to link prediction.
Inspired by convolutional neural networks (CNNs) [30] and network embeddings, GNNs were introduced to address the limitations of traditional embedding methods [9]. While CNNs are designed for Euclidean data, such as images and text, their performance is typically unfavourable in non-Euclidean data structures - a prevalence in complex networks. Moreover, despite their contributions to network embeddings, encoding methods such as DeepWalk and Node2Vec are constrained by their shallow learning mechanisms, which limit further improvements in network embedding quality. Appropriately, GNNs were developed to mitigate these issues [31].
Graph convolutional networks (GCNs) [32] are an effective adaptation of CNNs, and are designed for semi-supervised learning on graph data. GCNs learn hidden layer representations that incorporate both local network structures and node features without requiring a comprehensively labelled data set (Hamilton, Graph representation learning, 2020). Graph sample and aggregation (GraphSAGE) [33] is another influential GNN model that enables inductive learning by generating embeddings for unseen nodes during training. Unlike transductive models such as GCNs, GraphSAGE does not operate on the entire graph during training. Instead, the model learns to sample and aggregate features from the local neighbourhood of a node (Hamilton W. L., Graph representation learning, 2020). Last, graph attention networks (GATs) introduce the powerful attention mechanism [34], which significantly enhances the adaptability of the model. Unlike previous GNN models that use uniform weights in the aggregation function, GAT assigns varying weights to different nodes in a neighbourhood, based on their relative importance [8, 35].
GNN-based embeddings for link prediction exhibit both strengths and weaknesses. GNNs excel in capturing complex structural patterns by leveraging rich connectivity information in graphs. In addition, GNN-based approaches are versatile, accommodating various graph properties and auxiliary features. Interpretability can be difficult, however, owing to the partial black-box nature of GNNs. Scalability is also a concern in the context of large-scale networks, as training and inference can be computationally demanding [9]. Addressing these challenges is crucial for the effective application of GNN-based embeddings in link prediction.
It should be noted that different network embedding methods (such as GNN or random walk-based approaches) adopt various modelling pipelines for link prediction. Some methods directly generate link probabilities, while other methods require additional learning that is based on the node embeddings [24, 36, 37]. Two prevalent approaches involve employing a measure of similarity between two node embeddings, such as the dot product (representing the link probability), or treating the problem as a binary classification task. The latter approach, deemed more effective by Gurukar et al. [38], requires the computation of node-pair embeddings prior to classification. Therefore, an operator is applied to obtain the node-pair representations, which are then provided as input to a binary classifier. The operator choice varies across studies, and is sometimes completely left out of the documentation [29, 39, 40].
3. METHODOLOGY
In this section, the methodology employed to carry out a comparative study of link prediction techniques is described. First, the selection criteria for link prediction methods are outlined. Thereafter, the data sets under consideration and the associated pre-processing techniques are discussed in more detail. Evaluation techniques are then detailed and, finally, the correlation analysis is clarified.
3.1. Selection of link prediction methods
In this paper a computational study of the prevalent link prediction methods in the literature is done, as discussed in Section 2. The following heuristic-based methods are considered: CN, JC, SI, AA, PA, and RA. In the case of classifier-based methods, the following supervised learning algorithms are considered: decision tree [41], random forest (Breinman, 2001), logistic regression [41], support vector machine [43], and multi-layered perceptron [44]. Embedding-based methods include DeepWalk, Node2Vec, GcN, GraphSAGE, and GAT. The aforementioned selection of algorithms is guided by both diversity (in respect of their fundamental working) and their varying performance levels, as reported in different studies [4, 45, 46].
It is reported that these various approaches can exploit different underlying signals of similarity - a finding that underscores the empirical prevalence of the 'no free lunch' theorem [47]. A comprehensive comparison between these methods has not been addressed, despite their apparent success in a wide range of empirical studies [5, 6, 11]. It should be noted that studies reporting on such comparisons frequently offer an incomplete performance evaluation, often neglecting the critical precision-recall measure. The inclusion of precision-recall analysis enriches the understanding of algorithmic performance, and facilitates a more informative comparison, especially in the binary classification context [13]. This shortcoming inhibits conclusive inferences about the relative merits of these methods, underscoring the need for further research that is grounded in more robust evaluations [10]. In addition, a large number of the empirical evaluations to date have been based on a limited test suite (or sample size) of network data sets and link prediction algorithms, which inhibits both a holistic understanding of algorithmic performance and comprehension of the nuances associated with the different approaches. Furthermore, the extent to which different methods (or methodological paradigms) capture similar underlying features for link prediction is not realised [45]. Appropriately, this study seeks to address these limitations in the literature by considering a diverse set of representative link prediction methods for various network data sets, from which insight can be drawn so as to aid algorithmic selection decision support.
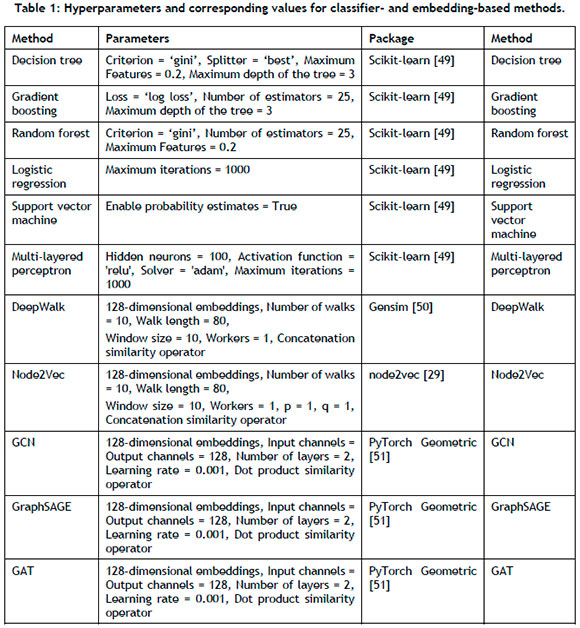
A notable scope limitation is required because of the computational time required to perform hyperparameter tuning. Consequently, this study employs the hyperparameter values delineated in the original papers of the respective methods. The authors do acknowledge, however, the importance of hyperparameter tuning, which can markedly affect link prediction performance, especially upon considering network variations across data sets. In Table 1, the implemented hyperparameter values and associated Python packages are detailed. The NetworKit [48] package was used for the heuristic-based link prediction methods. The experiments in this study were conducted on a MacBook Pro equipped with an Apple M1 chip and 8 GB of memory.
3.2. Selection of data sets
Surveys and frameworks reported in prior research predominantly focus on network data from a specific domain, such as social networks [6, 11, 22]. This data set selection bias may thus inadvertently obscure the perceived performance of a link prediction approach, inhibiting a more comprehensive evaluation of diverse problem contexts. To address this limitation, the proposed approach involves the inclusion of data sets from multiple domains, facilitating a more robust evaluation of the considered methods and their adaptability to different network types. The algorithmic performance comparison (and the insights inferred from it) can form the basis on which data-driven and domain-agnostic algorithmic selection can be performed. The adopted experimental design uses eight undirected, unweighted real-world networks obtained from diverse domains.
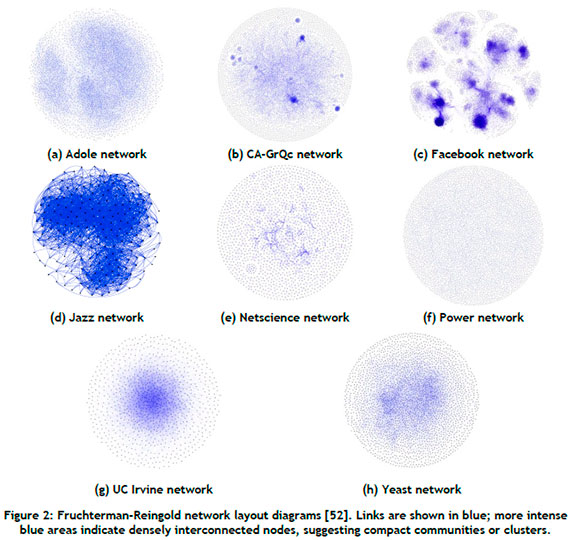
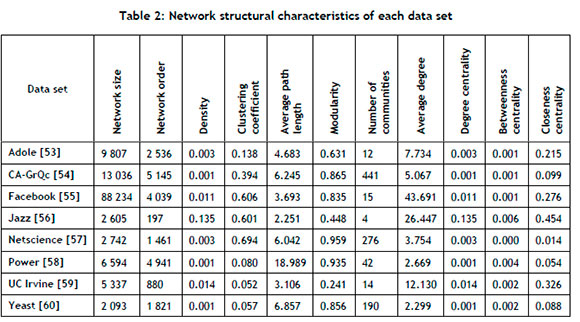
Visual representations showcasing the structural differences of the respective networks are presented in Figure 2. Furthermore, the numerical structural characteristics of each network data set are provided in Table 2. The Adole network is sparsely connected with significant modularity. The CA-GrQc network can be characterised by its high level of interconnectedness within its communities, but it is sparsely connected overall. A 'community' is defined as a subset of nodes within the network such that connections between the nodes are denser than connections with the rest of the network [25]. The Facebook network shows dense connections and a strong community structure. The Jazz network is similarly dense and interconnected, with a short average path length. A 'path' is a sequence of nodes connected by links, with its length being the number of traversed links [25]. The Netscience network has a robust community structure, but sparse overall connections. The Power network has distinct communities, but is not densely connected and has the longest average path length of all of the networks. The UC Irvine network has nodes with a high average degree and short average path length, but lacks a distinct community structure. The Yeast network is sparsely connected, but has a prominent community structure. It is assumed that no data quality issues are present in the benchmark data sets.
3.3. Description of data pre-processing techniques
During the data pre-processing phase of link prediction, careful consideration must be given to the nature of the input data. This involves splitting present links within the graph G = (V, £) into train and test sets, denoted by εtrain and εtest respectively, while, importantly, also generating missing links to include in these sets. Various sampling strategies can be implemented for the present links so as to split ε into εtrain and εtest, such as random sampling [38], spanning tree [61], and depth-first tree [37]. The choice of link sampling strategy can have an impact on the characteristics of the training data, and so can affect model performance by potentially excluding informative network features [37]. In this paper, a random sampling strategy is implemented that helps to mitigate the risk of introducing bias, ensuring a fair representation of the network's structure in the training data [10]. The computational simplicity of this strategy further warrants its selection.
Apart from partitioning present links, it is also important to generate missing links for inclusion in £train and εtest. However, a challenge relates to the significant imbalance between possible missing and present links [10, 13]. Various strategies have been proposed for effective sampling, such as random selection for broad coverage, although that approach may disregard structural characteristics and geodesic distance-based selection, which can introduce bias [3, 13, 62, 63]. Other approaches select links based on specific contextual information, such as constraining link selection to nodes having a degree of at least two [4]. A judicious combination of strategies is essential to sample effectively from the highly imbalanced set of candidates, ensuring balanced and representative train and test sets. In this study a hybrid approach is adopted, according to which the generation of missing links is (over)inflated by one order of magnitude when compared with the number of present links. Naturally, the intuitive approach is to counter the inherent imbalance. Preferential and geodesic sampling methods are integrated, according to which nodes with higher degrees are selected that reflect real-world network attachment preferences, while the geodesic method samples node pairs within a two-hop distance so as to capture local structure. (A 'hop' refers to the step or movement from one node to another via a single edge or link [25].) This approach aims to construct a diverse and representative set of negative samples, enhancing the robustness of link prediction evaluations. This adopted approach is, to the best of the authors' knowledge, novel. Data preprocessing was performed using a combination of the NetworKit [48] and NetworkX [64] Python packages.
The train-test split ratio varies widely in the literature, with Grover and Leskovec [29] employing a 50-50 split, Gao et al. [65] opting for 60-40, and Lai et al. [66] using 80-20. Such inconsistencies highlight the importance of careful consideration in preparing data sets for link prediction analysis. In this paper, an 8020 split is adopted, with under-sampling applied to the training set to address class imbalance. To verify the model, five-fold cross-validation is employed, offering robustness against overfitting.
Various strategies have been employed for feature selection in classifier-based link prediction, according to which some incorporate proximity, aggregated, and topological features, while others employ neighbour-based metrics, centrality metrics, community metrics, and geodesic distance [3, 22]. In this article, feature extraction is governed by standard network features such as CN, JC, and node degrees, chosen for their suitability for capturing network topology without domain-specific knowledge. In order to alleviate computational inefficiencies and reduce training times, the extracted scope features are delimited. This approach seeks to offer a succinct yet informative representation of the network structure for effective link prediction analysis.
3.4. Evaluation techniques
A large number of link prediction studies tend to rely heavily on reporting the area under the receiver operating characteristic curve (AUROC) as the primary performance metric, overlooking the importance of other evaluation measures [8, 11, 13, 67, 68]. Relying exclusively on AUROC may not provide a comprehensive evaluation in the context of link prediction. Integrating the area under the precision-recall (AUPR) curve with the commonly used AUROC is imperative for a more robust evaluation [8, 10, 13]. AUROC employs both the true positive rate and the false positive rate, providing a measure of a model's performance that is biased towards false positives (Hanley & McNeil, 1982). On the other hand, AUPR uses precision and recall, providing a measure of the model's efficacy in identifying positive samples. AUROC is suitable when positives and negatives are nearly balanced, whereas AUPR is favoured in cases of class imbalance or when differing costs are attached to false positives and negatives - typically the case for link prediction. AUROC can therefore be overly optimistic when applied to imbalanced data sets owing to the influence of true negatives, rendering AUPR more informative, as it gives the necessary attention to the minority class. In summary, employing both AUROC and AUPR yields a multifaceted and more accurate assessment of model performance in link prediction tasks, particularly in diverse conditions and data sets [13]. Appropriately, both AUPR and AUROC are employed as evaluation metrics.
3.5. Correlative investigation
A correlation analysis of network characteristics (summarised in Table 2) and algorithmic performance (i.e., AUPR) can yield valuable and actionable insight into algorithmic performance and offer decision support for algorithm selection. Appropriately, the Pearson's coefficient is employed. (The widely adopted Pearson's coefficient is a statistical measure employed to assess linear correlation between two variables [70].) The coefficient ranges from -1 to 1, according to which -1,1 and zero signify a perfectly negative, perfectly positive, and no linear correlation respectively. Values between -1 and 1 indicate varying strengths of negative or positive correlations.
It should be noted that previous research in the field has focused primarily on specific categories of link prediction method, such as heuristic-based approaches, or has restricted the analysis to specific network types, such as social networks [11, 12]. Furthermore, previous studies have typically examined the correlation between a single network characteristic and performance, primarily measured using AUROC [12]. The investigation presented in this paper is a more comprehensive approach to analysing correlative relationships between algorithmic performance (AUPR) and network characteristics (considering a wide range of features). The extent of this investigation is another novel contribution to the field of link prediction.
4. ANALYSIS
In this section, the findings stemming from the computational analyses that were carried out are presented. First, the algorithmic performance achieved by each of the link prediction methods is discussed, followed by a correlative analysis to discern the relationships between network characteristics and link prediction performance.
4.1. Algorithmic performance
The performance results of the heuristic-, classifier-, and embedding-based methods are illustrated graphically in Figure 3. A variety of noteworthy outcomes can be highlighted from examining the results, alluding to some preliminary insight into the qualitative extent to which specific network characteristics can affect the performance of link prediction methods. For example, the networks with large clustering coefficients, such as Facebook and Netscience, exhibited consistently favourable link prediction performance, as showcased by the high AUROC and AUPR scores across all categories of link prediction techniques; it can thus be conjectured that these techniques perform commendably in the context of highly clustered networks. Another noteworthy observation is the markedly strong performance of classifier-based methods, such as the random forest and the multi-layered perceptron, in respect of the Facebook data set, achieving nearly perfect AUROC and AUPR scores, thus reinforcing the notion of their utility in the context of networks that have notable clustering and relatively short average path lengths. In the case of the Power and Yeast networks (each having long average path lengths), the performance of heuristic- and classifier-based methods was less satisfactory, underlining their limited utility in the context of these specific types of networks.
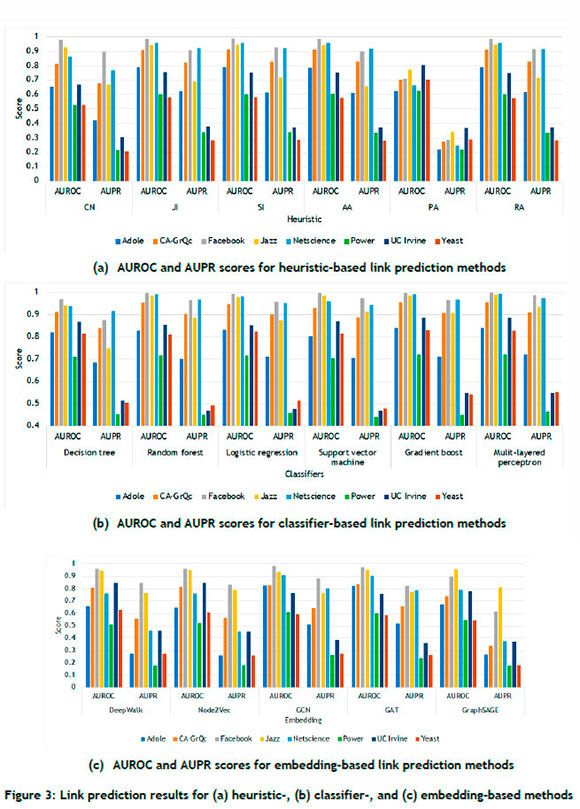
Conversely, embedding-based methods such as GCN and GAT exhibited relative superior performance in the case of networks with high modularity, such as CA-GrQc and Power. Their performance did show, however, a noticeable decline in respect of the Adole and Power networks, which had smaller clustering coefficients, indicating an inherent limitation of these methods when applied to less clustered networks. The discrepancy between the AUROC and AUPR results, particularly evident in the case of the Power and UC Irvine networks, substantiates the assertion that, while AUROC is sensitive to true positive rate, it does not account sufficiently for the true negatives, rendering AUPR a more informative measure. Consequently, the evaluation of prediction methods needs a careful consideration of both metrics in tandem with network characteristics so as to ensure a robust and accurate assessment.
4.2. Correlative analysis
Correlations between various network characteristics and the algorithmic performance of the link prediction methods are now examined. Attention is particularly devoted to the top six network characteristics in respect of the largest correlation magnitude; this delimitation facilitates a more focused understanding of the main insights that can be inferred. Performance is assessed exclusively in respect of AUPR owing to its stated superiority for link prediction problems. It should be noted that the averaged network characteristic values and AUPR scores (across all data sets) were considered in this analysis.
A summary of the results from the correlative analysis is presented in Figure 4. It can be observed that the clustering coefficient has notably strong positive correlations with the performance across the majority of link prediction methods. This indicates that these algorithms efficiently exploit the community structures that are often reflected by high clustering coefficients. However, modularity reveals less definitive patterns in respect of correlations, which is indicative of a more nuanced relationship. One can therefore suggest that a network's modularity should be carefully considered before attempting to select an appropriate algorithmic approach. The average degree shows a wide range of positive correlations, especially with respect to node-embedding. The average degree shows a wide range of positive correlations, especially with respect to node-embedding methods such as DeepWalk and Node2Vec. This may be attributed to the notion that a higher average degree generally corresponds to a richness in structural information that can be exploited by these methods.
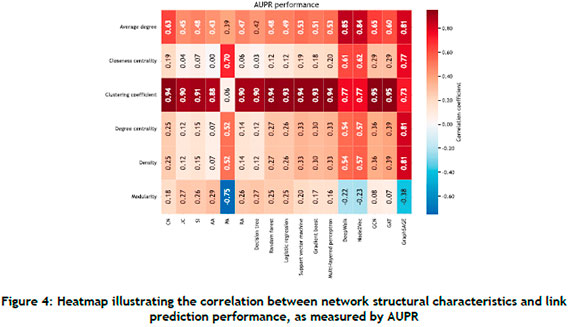
Differences in correlations across various methods are noted. For instance, heuristic methods such as CN and JC show a distinct pattern when compared with embedding-based approaches such as DeepWalk and Node2Vec. This distinction demonstrates that the influence of network characteristics on performance can vary substantially, depending on the applied algorithmic approach.
5. CONCLUSION
Based on the insights inferred from the analyses above, it is evident that careful consideration should be given to the strengths, limitations, and applicability of various link prediction methods. Experimental comparisons highlighted the pivotal role of network characteristics in prediction performance. Accordingly, densely connected and community-structured networks (exemplified by Facebook and Netscience) showed admirable link prediction performance across various methods. Conversely, sparsely connected networks with longer average path lengths (such as the Power and Yeast networks) posed considerable prediction difficulties. Among the techniques considered in this study, classifier-based methods - in particular, sophisticated models such as multi-layered perceptrons - emerged as superior in performance, adeptly addressing intricate network patterns computationally. Heuristic methods displayed a commendable performance, while the success of embedding-based methods relied on network density and community structure.
Additional quantitative-based insights were inferred from the correlative analysis that was carried out, which demonstrated varied relationships between network properties and prediction performance. Notably, a strong positive correlation was observed between the clustering coefficient and the prediction performance, while average path length showed a moderately negative correlation.
For future work, expanding on the test suite by incorporating synthetic data sets into the evaluation is recommended. Including synthetic data sets could both facilitate a more comprehensive analysis and enable an investigation into algorithm performance under varying (predefined) conditions, thereby enhancing the value of the conclusions that are drawn. In addition, developing a structured and automated algorithm selection framework for link prediction algorithms, based on network characteristics, could streamline the process of choosing the best-suited method, improving efficacy and reducing the risk of selection bias.
In conclusion, the results from the study have helped to provide an improved understanding of link prediction methods via the comprehensive comparative study carried out in this paper. The importance of considering network characteristics in method selection is substantiated by the empirical findings. This aforementioned is exemplified by the superior accuracy of classifier-based methods and the competitive performance of embedding-based techniques in certain problem contexts. Consequently, the findings in this study represent a contribute to the field of link prediction by proffering valuable and actionable insight into respect of understanding algorithmic performance and by providing a basis on which decision support for algorithm selection can be pursued.
REFERENCES
[1] T. G. Lewis, Network science: Theory and applications, Hoboken, NJ: John Wiley & Sons, 2011. [ Links ]
[2] M. E. J. Newman, Networks, New York, NY: Oxford University Press, 2018. [ Links ]
[3] M. Al-Hasan, V. Chaoji, S. Salem and M. Zaki, "Link prediction using supervised learning," in Workshop on Link Analysis, Counterterrorism and Security, Newport Beach, Ca, 2005. [ Links ]
[4] D. Liben-Nowell and J. Kleinberg, "The link prediction problem for social networks," in Proc. 12th CIKM International Conference on Information and Knowledge Management, New Orleans, LA, 2003. [ Links ]
[5] Y. Yang, R. N. Lichtenwalter and N. V. Chawla, "Evaluating link prediction methods," Knowledge and Information Systems, vol. 45, no. 1, pp. 751-782, 2015. [ Links ]
[6] P. Wang, B. Xu, Y. Wu and X. Zhou, "Link prediction in social networks: The state-of-the-art," Science China Information Sciences, vol. 58, no. 1, pp. 1-38, 2015. [ Links ]
[7] A. Kumar, S. S. Singh, K. Singh and B. Biswas, "Link prediction techniques, applications, and performance: A survey," Physica A: Statistical Mechanics and its Applications, vol. 553, no. 1, p. 124289, 2020. [ Links ]
[8] H. Wu, C. Song, Y. Ge and T. Ge, "Link prediction on complex networks: An experimental survey," Data Science and Engineering, vol. 7, no. 3, pp. 253-278, 2022. [ Links ]
[9] M. Zhang and Y. Chen, "Link prediction based on graph neural networks," in Proc. 32nd Conference on Neural Information Processing Systems, Montréal, 2018. [ Links ]
[10] R. N. Lichtenwalter and N. V. Chawla, "Link prediction: Fair and effective evaluation," in Proc. 18th International Conference on Advances in Social Networks Analysis and Mining, Istanbul, 2012. [ Links ]
[11] F. Gao, K. Musial, C. Cooper and S. Tsoka, "Link prediction methods and their accuracy for different social networks and network metrics," Scientific Programming, vol. 2015, pp. 1-13, 2015. [ Links ]
[12] X. Feng, J. Zhao and K. Xu, "Link prediction in complex networks: A clustering perspective," The European Physical Journal B, vol. 85, no. 1, pp. 1-9, 2012. [ Links ]
[13] R. N. Lichtenwalter, J. T. Lussier and N. V. Chawla, "New perspectives and methods in link prediction," in Proc. 16th International Conference on Knowledge Discovery and Data Mining, Washington, DC, 2010. [ Links ]
[14] E. Sprinzak, S. Sattath and H. Margalit, "How reliable are experimental protein-protein interaction data?,", Journal of Molecular Biology, vol. 327, no. 5, pp. 919-923, 2003. [ Links ]
[15] A. Szilagyi, V. Grimm, A. K. Arakaki and J. Skolnick, "Prediction of physical protein-protein interactions," Physical Biology, vol. 2, no. 2, pp. 1-16, 2005. [ Links ]
[16] M. E. J. Newman, "Clustering and preferential attachment in growing networks," Physical Review E, vol. 64, no. 2, p. 025102, 2001. [ Links ]
[17] P. Jaccard, "The distribution of the flora in the alpine zone," New Phytologist, vol. 11, no. 2, pp. 241-272, 1912. [ Links ]
[18] G. Salton, Introduction to modern information retrieval, New York, NY: McGraw-Hill, 1983. [ Links ]
[19] L. A. Adamic and E. Adar, "Friends and neighbors on the web," Social Networks, vol. 25, no. 3, pp. 211-230, 2003. [ Links ]
[20] A. L. Barabási, H. Jeong, Z. Néda, E. Ravasz, A. Schubert and T. Vicsek, "Evolution of the social network of scientific collaborations," Physica A: Statistical Mechanics and its Applications, vol. 311, no. 4, pp. 590-614, 2002. [ Links ]
[21] T. Zhou, L. Lü and Y. Zhang, "Predicting missing links via local information," The European Physical Journal B, vol. 71, pp. 623-630, 2009. [ Links ]
[22] S. A. Curiskis, T. R. Osborn and P. J. Kennedy, "Link prediction and topological feature importance in social networks," in Proc. 13th Australasian Data Mining Conference, Sydney, 2015. [ Links ]
[23] P. Cui, X. Wang, J. Pei and W. Zhu, "A survey on network embedding," Transactions on Knowledge and Data Engineering, vol. 31, no. 5, pp. 833-852, 2018. [ Links ]
[24] D. Zhang, J. Yin, X. Zhu and C. Zhang, "Network representation learning: A survey," Transactions on Big Data, vol. 6, no. 1, pp. 3-28, 2020. [ Links ]
[25] M. A. Henning and J. H. van Vuuren, Graph and network theory: An applied approach using Mathematica, Cham: Springer, 2022. [ Links ]
[26] B. Perozzi, R. Al-Rfou and S. Skiena, "DeepWalk: Online learning of social representations," in Proc. 20th International Conference on Knowledge Discovery and Data Mining, New York, NY, 2014. [ Links ]
[27] T. Mikolov, K. Chen, G. Corrado and J. Dean, "Efficient estimation of word representations in vector space," in Proc. 1st International Conference on Learning Representation, Scottsdale, AZ, 2013. [ Links ]
[28] W. L. Hamilton, "Graph representation learning," Synthesis Lectures on Artificial Intelligence and Machine Learning, vol. 14, no. 3, pp. 1-159, 2020. [ Links ]
[29] A. Grover and J. Leskovec, "Node2Vec: Scalable feature learning for networks," in Proc. 22nd International Conference on Knowledge Discovery and Data Mining, San Franciso, CA, 2016. [ Links ]
[30] P. Sermanet, S. Chintala and Y. LeCun, "Convolutional neural networks applied to house numbers digit classification," in Proc. 21st International Conference on Pattern Recognition, Tsukuba, 2012. [ Links ]
[31] J. Zhou, G. Cui, S. Hu, Z. Zhang, C. Yang, Z. Liu, L. Wang, C. Li and M. Sun, "Graph neural networks: A review of methods and applications," AI Open, vol. 1, pp. 57-81, 2020. [ Links ]
[32] T. N. Kipf and M. Welling, "Semi-supervised classification with graph convolutional networks," in Proc. 5th International Conference on Learning Representations, Toulon, 2017. [ Links ]
[33] W. Hamilton, Z. Ying and J. Leskovec, "Inductive representation learning on large graphs," in Proc. 31st Conference on Neural Information Processing Systems, Long Beach, CA, 2017. [ Links ]
[34] A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser and I. Polosukhin, "Attention is all you need," in Proc. 31st Annual Conference on Neural Information Processing Systems, Long Beach, CA, 2017. [ Links ]
[35] P. Velivckovic, G. Cucurull, A. Casanova, A. Romero, P. Lio and Y. Bengio, "Graph attention networks," in Proc. 6th International Conference on Learning Representations, Vancouver, 2018. [ Links ]
[36] B. Kang, J. Lijffijt and T. de Bie, "Conditional network embeddings," in Proc. 7th International Conference on Learning Representations, New Orleans, LA, 2019. [ Links ]
[37] A. Mara, J. Lijffijt and T. de Bie, "Benchmarking network embedding models for link prediction: Are we making progress?," in Proc. 7th International Conference on Data Science and Advanced Analytics, Sydney, 2020. [ Links ]
[38] S. Gurukar, P. Vijayan, A. Srinivasan, G. Bajaj, C. Cai, M. Keymanesh, S. Kumar, P. Maneriker, A. Mitra and V. Patel, B Ravindran, S Parthasarathy Network representation learning: Consolidation and renewed bearing, 2019. [ Links ]
[39] A. Tsitsulin, D. Mottin, P. Karras and E. Müller, "Verse: Versatile graph embeddings from similarity measures," in Proc. 27th International World Wide Web Conference, Lyon, 2018. [ Links ]
[40] D. Wang, P. Cui and W. Zhu, "Structural deep network embedding," in Proc. 22nd International Conference on Knowledge Discovery and Data Mining, Washington, DC, 2016. [ Links ]
[41] L. Breiman, Classification and regression trees, New York, NY: Routledge, 1984. [ Links ]
[42] L. Breiman, "Random forests," Machine Learning, vol. 45, pp. 5-32, 2001. [ Links ]
[43] B. E. Boser, I. M. Guyon and V. N. Vapnik, "A training algorithm for optimal margin classifiers," in Proc. 5th Annual Workshop on Computational Learning Theory, Pittburgh, PA, 1992. [ Links ]
[44] F. Rosenblatt, "The perceptron: A probabilistic model for information storage and organization in the brain," Psychological Review, vol. 65, no. 6, p. 386-408, 1958. [ Links ]
[45] A. Ghasemian, H. Hosseinmardi, A. Galstyan, E. M. Airoldi and A. Clauset, "Stacking models for nearly optimal link prediction in complex networks," Proceedings of the National Academy of Sciences, vol. 117, no. 38, pp. 23393-23400, 2020. [ Links ]
[46] A. Ghasemian, H. Hosseinmardi and A. Clauset, "Evaluating overfit and underfit in models of network community structure," IEEE Transactions on Knowledge and Data Engineering, vol. 32, no. 9, pp. 1722-1735, 2019. [ Links ]
[47] D. H. Wolpert and W. G. Macready, "No free lunch theorems for optimization," IEEE Transactions on Evolutionary Computation, vol. 1, no. 1, pp. 67-82, 1997. [ Links ]
[48] E. Angriman, A. van der Grinten, M. Hamann, H. Meyerhenke and M. Penschuck, "Algorithms for large-scale network analysis and the NetworKit toolkit," in Bast, H., Korzen, C., Meyer, U., Penschuck, M. (eds), Algorithms for big data, lecture notes in computer science, vol. 13201, Cham: Springer, 2023, pp. 3-20. [ Links ]
[49] F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot and E. Duchesnay, "Scikit-learn: Machine learning in Python," Journal of Machine Learning Research, vol. 12, pp. 2825-2830, 2011. [ Links ]
[50] R. Rehurek and P. Sojka, "Software framework for topic modelling with large corpora," in Proc. LREC Workshop on New Challenges for NLP Frameworks, Valletta, Malta, 2010. [ Links ]
[51] M. Fey and J. E. Lenssen, "Fast graph representation learning with PyTorch Geometric," in Proc. 2nd ICLR Workshop on Representation Learning on Graphs and Manifolds, New Orleans, LA, 2019. [ Links ]
[52] T. M. J. Fruchterman and E. M. Reingold, "Graph drawing by force-directed placement," Software: Practice and Experience, vol. 21, no. 11, pp. 1129-1164, 1991. [ Links ]
[53] J. Moody, "Peer influence groups: Identifying dense clusters in large networks," Social Networks, vol. 23, no. 4, pp. 261-283, 2001. [ Links ]
[54] M. E. J. Newman, "The structure of scientific collaboration networks," National Academy of Sciences, vol. 98, no. 2, pp. 404-409, 2001. [ Links ]
[55] J. Leskovec and A. Krevl, SNAP datasets: Stanford large network dataset collection, 2014. [ Links ]
[56] P. M. Gleiser and L. Danon, "Community structure in Jazz," Advances in Complex Systems, vol. 6, no. 4, pp. 565-573, 2003. [ Links ]
[57] M. E. J. Newman, "Finding community structure in networks using the eigenvectors of matrices," Physical Review E, vol. 74, no. 3, p. 036104, 2006. [ Links ]
[58] D. J. Watts and S. H. Strogatz, "Collective dynamics of 'small-world' networks," Nature, vol. 393, no. 6684, pp. 440-442, 1998. [ Links ]
[59] T. Opsahl, "Triadic closure in two-mode networks: Redefining the global and local clustering coefficients," Social Networks, vol. 35, no. 2, pp. 159-167, 2013. [ Links ]
[60] J. D. J. Han, D. Dupuy, N. Bertin, M. E. Cusick and M. Vidal, "Effect of sampling on topology predictions of protein-protein interactions," Nature Biotechnology, vol. 23, no. 7, pp. 839-844, 2005. [ Links ]
[61] A. Mara, J. Lijffijt and T. de Bie, "Evalne: A framework for evaluating network embeddings on link prediction," Software X, vol. 17, p. 100997, 2022. [ Links ]
[62] C. Wang, V. Satuluri and S. Parthasarathy, "Local probabilistic models for link prediction," in Proc. 7th IEEE International Conference on Data Mining, Omaha, NE, 2007. [ Links ]
[63] S. Scellato, C. Mascolo, M. Musolesi and V. Latora, "Distance matters: Geo-social metrics for online social networks," in 3rd Conference on Online Social Networks, Boston, MA, 2010. [ Links ]
[64] A. Hagberg, P. Swart and D. S. Chult, "Exploring network structure, dynamics, and function using NetworkX," in Proceedings of the 7th Python in Science Conference (SciPy 2008), G. Varoquaux, T. Vaught, J. Millman (Eds), 2008, pp. 11-15. [ Links ]
[65] M. Gao, L. Chen, X. He and A. Zhou, "Bine: Bipartite network embedding," in Proc. 41st International Conference on Research & Development in Information Retrieval, New Brunswick, NJ, 2018. [ Links ]
[66] Y. Lai, C. Hsu, W. H. Chen, M. Yeh and S. Lin, "Prune: Preserving proximity and global ranking for network embedding," Advances in Neural Information Processing Systems, vol. 30, no. 1, pp. 52575266, 2017. [ Links ]
[67] L. Lü, C. Jin and T. Zhou, "Similarity index based on local paths for link prediction of complex networks," Physical Review E, vol. 80, no. 4, p. 046122, 2009. [ Links ]
[68] W. Cukierski, B. Hamner and B. Yang, "Graph-based features for supervised link prediction," in Proc. IEEE International Joint Conference on Neural Networks , San Jose, CA, 2011. [ Links ]
[69] J. A. Hanley and B. J. McNeil, "The meaning and use of the area under a receiver operating characteristic (ROC) curve," Radiology, vol. 143, no. 1, pp. 29-36, 1982. [ Links ]
[70] J. L. Rodgers and A. W. Nicewander, "Thirteen ways to look at the correlation coefficient," The American Statistician, vol. 42, no. 1, pp. 59-66, 1988. [ Links ]
[71] L. Katz, "A new status index derived from sociometric analysis," Psychometrika, vol. 18, no. 1, pp. 39-43, 1953. [ Links ]
* Corresponding author: Brown.lienke@gmail.com
ORCID® identifiers
L.M. Brown: https://orcid.org/0000-0001-5361-4688
G.S. Nel: https://orcid.org/0000-0002-0293-1234














