Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
African Journal of Primary Health Care & Family Medicine
On-line version ISSN 2071-2936
Print version ISSN 2071-2928
Afr. j. prim. health care fam. med. (Online) vol.12 n.1 Cape Town 2020
http://dx.doi.org/10.4102/phcfm.v12i1.2264
ORIGINAL RESEARCH
Geographical distribution and antibiotics susceptibility patterns of toxigenic Vibrio cholerae isolates from Kisumu County, Kenya
Silas O. AwuorI; Eric O. OmwengaI; Ibrahim I. DaudII
IDepartment of Health, School of Health Sciences, Kisii University, Kisii, Kenya
IIKenya Medical Research Institute, United States Army Medical Research Directorate-Africa, HJF Medical Research International, Kericho, Kenya
ABSTRACT
BACKGROUND: Multiple drug resistance has become a major threat to the treatment of cholera. Recent studies in Kenya have described the epidemiology, especially the risk factors, of cholera; however, there is little information on the phenotypic and drug susceptibility patterns of Vibrio cholerae (V. cholerae) in outbreaks that in the recent past have occurred in western Kenya.
AIM: To characterise and determine the antibiotics' susceptibility profiling of toxigenic V. cholerae isolates from Kisumu County.
SETTING: The project was conducted in Kisumu County, Kenya.
METHODS: A total of 119 V. cholerae O1, biotype El Tor, isolates collected during 2017 cholera outbreak in Kisumu County were used for this study. The samples were cultured on thiosulphate-citrate-bile salts sucrose (TCBS) agar and biochemical tests were carried out using standard procedures. Susceptibility tests were conducted by using various conventional antibiotics against standard procedures.
RESULTS: Of the 119 isolates, 101 were confirmed to be V. cholerae belonging to serotypes Inaba and Ogawa, with Inaba being the predominant serotype (73.95%). The isolates were susceptible to ciprofloxacin (100%), ofloxacin (100%), gentamycin (100%), doxycycline (99%), ceftriaxone (99%) and streptomycin (96.04%) antimicrobials, and resistant to erythromycin (53.47%), amoxicillin (64.4%), nalidixic acid (83.2%) and ampicillin (89.11%), with high resistance to cotrimoxazole (99%) and tetracycline (97%).
CONCLUSION: Vibrio cholerae was resistant to multiple antibiotics, including those commonly used in the management of cholera. Taken together, there is a need to carry out regular surveillance on antimicrobial drug resistance during outbreaks.
Keywords: Vibrio cholerae; antimicrobial susceptibility; Kisumu; tetracycline; cotrimoxazole; Kenya.
Introduction
Antibiotic resistance has been increasing since the introduction of antibiotics in the middle of the 20th century.1,2 Since the introduction of tetracyclines, many other antibiotics have been introduced, but each has shown resistance at some level.3,4 The level of circulating virulent, multiple drug-resistant (MDR) bacteria has been a global threat to healthcare.5,6 Toxigenic Vibrio cholerae (V. cholerae) O1 or O139 has been a cause of gastrointestinal infections and is involved in severe outbreaks of dehydrating diarrhoea in most of developing nations worldwide.1,2 Cholera - a disease associated with poor sanitation - is often transmitted by consumption of food and water contaminated with bacterium.7 Cholera re-emerged as a major infectious disease in the past, with a global increase in its incidence.8 In addition, there was unprecedented appearance of an epidemic strain of V. cholerae non-O1, classified as V. cholerae O139, in late 1992 in West Bengal state of India.9 Since 2007, Kenya has experienced cholera outbreaks characterised by increased mortality.10 For instance, a cholera outbreak in November 2007 that began in Nyanza claimed 67 lives out of 1243 cases by April 2008.11,12 Previous cholera outbreaks in Nyanza were not as severe as that of 2007.13 Currently, V. cholerae antimicrobial resistance has become a global concern as fewer new antibiotics are being discovered, with more pathogens becoming resistant to most commonly used antibiotics.14 Antibiotics that have been used in the management of cholera cases for decades such as tetracyclines, which include doxycycline and tetracycline, have been associated with the presence of IncC conjugative plasmids.15,16,17 In Kenya, doxycycline and tetracycline are the first-line drugs used for cholera treatment in adults. Erythromycin and chloramphenicol are used for the treatment of cholera in children and pregnant women.18 Despite the body of research on the epidemiology of cholera and its associated morbidity in Kenya, and Kisumu County in particular, studies focusing on the phenotypic and molecular characteristics of V. cholerae in outbreaks amongst Kisumu patients are limited and often anecdotal. This study is aimed to characterise and determine the antibiotics susceptibility profiling of toxigenic V. cholerae isolates from the various sub-counties of Kisumu County.
Materials and methods
Study design
This was a descriptive cross-sectional study that focussed on the 2017 cholera outbreak previously isolated from diarrhoeal stool specimens. The specimens were obtained from patients presenting with passage of three or more watery stools with or without vomiting during cholera outbreaks.
Study area
The study was performed at the six sub-counties of Kisumu County, including Muhoroni, Nyando, Nyakach, Kisumu West, Kisumu East and Seme.
Study samples
A total of 119 isolates of V. Cholerae O1, previously isolated from diarrhoeal stool specimens between January and December 2017 within the county, were used for this study. The isolates were stabilised and stored at -80 °C freezing condition at the Centre for Global Health Research.
Study procedure
Identification and susceptibility testing of Vibrio cholerae isolates
The isolates which were stored at -80 °C were revived under sterile conditions as described previously.17 The isolates were sub-cultured in thiosulphate-citrate-bile salts sucrose (TCBS) agar, which was prepared as per the manufacturer's instructions. The plates were then incubated at 37 °C for 18-24 h in aerobic conditions. After 24 h of incubation, bacterial cultures giving growth of pure green and yellow colonies were presumed to be that of V. cholerae and subjected to Gram staining, and biochemical and serological identification testing.
Gram staining
Thin smear was prepared from the colonies grown on TCBS media, air-dried and heat-fixed. The smear was then stained using Gram staining technique.19
Biochemical identification
The isolates were subjected to biochemical identification using Application Programming Interface (API) 20 E kit as per the manufacturer's instructions (API 20 E; BioMerieux, Charbonnieres-Les-Bains, France) together with established protocols.20,21,22,23 Briefly, 2 mL of API 0.85% NaCl was inoculated with pure colonies of young cultures (18-24-h old) to make a 0.5 McFarland standard and measured with the ATB densitometer (Denka Seiken Co., Ltd, Tokyo, Japan). Each cupule of the strip was dispensed with 56 microliter (µL) of suspension using ATB electronic pipette. The Arginine dihydrolase (ADH) Lysine decarboxylase (LDH) urea test (UREA) Arabinose fermentation (LARL) Ornithine dehydrogenase (ODC) Ketogluconate (KG) and 5KG tests were covered with two drops of mineral oil. The lid was placed on the strip and incubated at 36 °C ± 2 °C for 24 h (± 2 h) in aerobic conditions. One drop of JAMES reagent was added in IND reaction and the strips were read using mini API machine (BioMerieux, Charbonnieres-Les-Bains, France). The results were again read through computer and interpreted using mini API identification software (BioMerieux, France). All tests were performed in triplicate and were independent of each other.
Serological identification
The serologic identification was conducted by the slide agglutination technique with polyvalent anti-sera for V. cholerae O1 and O139, and monovalents for serotypes Inaba and Ogawa (Denka Seiken Co. Ltd., Japan) according to the manufacturer's instructions and previously published protocols.20,21,22,23 The confirmed V. cholerae isolates were further sub-cultured on nutrient agar (that was prepared as per the manufacturer's instructions) and incubated overnight at 37 °C for 18-24 h in aerobic conditions. A pure colony was picked aseptically and mixed in a drop of sterile normal saline on a glass slide, making a milky suspension. A drop of antiserum was added onto the drop and mixed. Agglutination reaction was observed. Positive agglutination confirmed serotype and subtype of isolates. In this test, K3 and K14 strains belonging to V. cholerae O1 biotype El Tor, subtype Ogawa, were used as positive controls, whilst Escherichia coli strain ATCC 25922 was used as a negative control. All tests were carried out in triplicate independent of one another.
Antimicrobial susceptibility testing
Susceptibility to antimicrobial agents was assayed by the disk diffusion method described previously.20 Drug incorporated disks were used for O/129 vibriostatic agent whereas, for the rest of the antimicrobials, their minimum inhibitory concentration (MIC) was determined by the Etest (previously known as Epsilometer test) method as used before.21Escherichia coli standard strain, ATCC 25922, was used as internal quality control. The procedure for antimicrobial susceptibility testing was performed as per the Clinical Laboratory Standards Institute (CLSI) guidelines.19 All tests were carried out in triplicate independent of one another.
Data analysis
All experiments were conducted in triplicate to validate reproducibility. Statistical analysis was performed using Stata software. Data on socio-demographics were summarised by frequencies and percentage values. All values of diameter zones of inhibition were reported as mean ± standard error. Data analysis was performed during and after collection. The study did not involve patients.
Ethical consideration
Confidentiality and privacy were strictly adhered to and no names of individuals were recorded or made known in the collection or reporting of information. The study was granted ethical clearance by the Board of Postgraduate Studies (BPS) of Kisii University, and ethical approval to conduct the study was sought from the Institutional Research Ethics Committee (IREC) at Moi University/Moi Teaching and Referral Hospital (MTRH) and the National Commission of Science, Technology and Innovations (NACOSTI).
Results
Socio-demographic data of the Vibrio cholerae cases in the 2017 cholera outbreaks in Kisumu
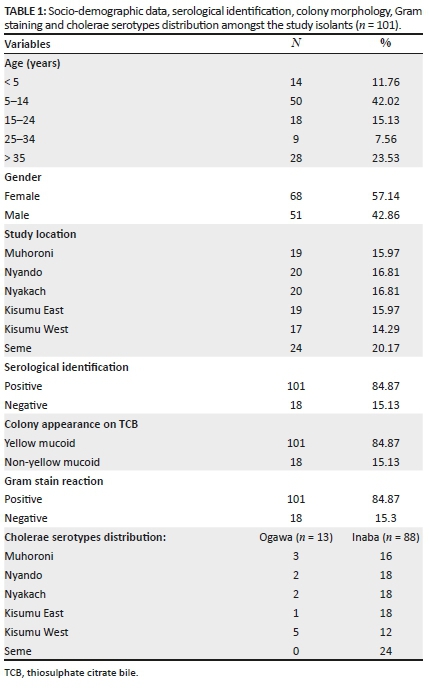
Samples were obtained from patients presenting with symptoms of cholera. The mean age of the patients was 22.62 years (range: 2-73 years). The most infected age group was 5-14 years (42%), and the least infected age group was 25-34 years. In terms of gender, women (57%) were more infected than men. Geographically, the majority of cases were from Seme sub-county (20%), whilst the least (14%) were from Kisumu West sub-county (Table 1).
Morphological and biochemical characteristics of isolated Vibrio cholerae
Of the 119 isolates, 101 (85%) were found positive for V. cholerae based on a combination of serological, biochemical and cultural identification techniques (Table 1). Growth characteristics of presumptive V. cholerae were determined on TCBS selective media as large yellow mucoid colonies, which was suggestive of the presence of V. cholerae strains. The colonies also produced Gram-negative rods by Gram staining (Table 1).
Next, all presumptive colonies of V. cholerae were subjected to further identification based on biochemical reactions. Isolates that were Gram-negative rods, positive for Voges-Proskauer and Indole tests, and negative for hydrogen sulphide test, together with glucose, sucrose and mannitol fermenters, were confirmed as V. cholera (Table 2).
Moreover, the presumptive V. cholerae isolates were further tested for agglutination with polyvalent O antisera and only positive isolates (84.9%) were stocked as V. cholerae.
Characterisation of Vibrio cholerae isolates
Biochemical identification using mini-API ID 20E (Biomeriux SA, France) confirmed that 101 (84.9%) isolates were V. cholerae isolates, whilst 18 (15.1%) were non-V. cholerae isolates. Serological identification further showed that 101 (84.9%) isolates were of V. cholerae O1. This test further indicated that both serotypes were present, with 88 (73.95%) serotypes being Inaba and 13 (10.92%) as serotype Ogawa (Table 1).
Distribution of cholera and cholera subtypes in the study sites
The majority of infected persons were from Seme sub-county (20.2%), followed by 16.8% from Nyando sub-county, 16.8% from Nyakach sub-county, 16.0% from Muhoroni Sub- county, 16.0% from Kisumu East sub-county, whilst the least (14.3%) were from Kisumu West sub-county. The distribution of Inaba and Ogawa between the study sites varied, with Kisumu West sub-county having the majority of serotypes Ogawa (29.41%) and Inaba (70.59%) (Table 1).
Antimicrobial susceptibility patterns
The positive isolates for V. cholerae were screened for their susceptibility to various antibiotics commonly used to manage cholera cases. All 100% isolates were susceptible to gentamicin at a concentration of 0.016 µg/mL - 256 µg/mL and ofloxacin at a concentration of 0.002 µg/mL - 32 µg/mL. However, there was a decreased susceptibility to chloramphenicol (55.45%) at a concentration of 0.016 µg/mL - 256 µg/mL. In all, 10.89% of the V. cholerae isolates were susceptible to ampicillin at a concentration of 0.016 µg/mL - 256 µg/mL, 99% to doxycycline at a concentration of 0.016 µg/mL - 256 µg/mL, 96.04% to streptomycin at a concentration of 0.064 µg/mL - 1024 µg/mL and 2.97% to tetracycline at a concentration of 0.016 µg/mL - 256 µg/mL. It was observed that 64.36% of V. cholerae isolates were resistant to amoxicillin at a concentration of 0.016 µg/mL - 256 µg/mL, 98.02% to cotrimoxazole at a concentration of 0.016 µg/mL - 256 µg/mL, 53.47% to erythromycin at a concentration of 0.016 µg/mL - 256 µg/mL, 97.03% to tetracycline at a concentration of 0.016 µg/mL - 256 µg/mL and 83.17% to nalidixic acid at a concentration of 0.016 µg/mL - 256 µg/mL. Escherichia coli ATCC 25922 showed susceptibility to all antibiotics used in this study at the above-mentioned concentrations (Table 3).
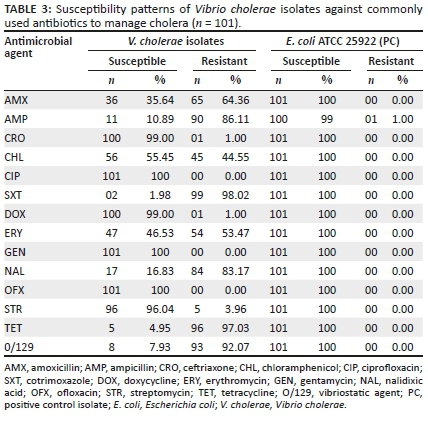
For Etest, all isolates were susceptible to ciprofloxacin and gentamicin. However, there was a reduced susceptibility to chloramphenicol, with 55.45% of isolates being susceptible and 4.55% were being resistant. The MIC ranged from 0.25 µg/mL to 96 µg/mL. In all, 99% of V. cholerae isolates were susceptible to ceftriaxone, 96.04% were susceptible to streptomycin and 99% were susceptible to doxycycline. The MIC50 of ceftriaxone, doxycycline and streptomycin were in the susceptible zone.
It was also deduced that 64.36% of the V. cholerae isolates were resistant to amoxicillin (MIC 0.75: > 256 µg/mL), 53.47% to erythromycin (MIC 0.125: 48 µg/mL) and 83.17% to nalidixic acid (MIC 0.125: > 256 µg/mL). The MIC50 of nalidixic acid, amoxicillin and erythromycin were in the resistant zone. These results are in agreement with the results obtained by the disk diffusion method. MIC50 for E. coli standard strain ATCC 25922 was susceptible to all antibiotics used in this study.
Susceptibility to O/129 vibriostatic agent
The evaluation of susceptibility towards the O/129 vibriostatic agent shows that the majority of the V. cholerae isolates (93 out of 101; 92.07%) were resistant to this agent at a concentration of 150 µg, with only 8 (7.93%) being sensitive with an inhibition of ≥ 16 mm (Table 3).
Discussion
Serological characterisation revealed that the majority of isolates (73.95%) belonged to serotype Inaba. This finding concurs with a previous study conducted on V. cholerae O1 strains isolated in the coastal region of Kenya between 2005 and 2007, where serotype Inaba emerged as the main cause of epidemic.16 Similar findings were reported in another study carried out on V. cholerae O1 strains isolated in Kisumu county in 2014, where serotype Inaba emerged as the main cause of epidemic. These findings suggest a shift in the occurrence of Ogawa and Inaba serotypes in a given area, which is thought to be a consequence of genetic reversal that makes it possible for the Ogawa strain, which was dormant in the environment, to be able to mutate to serotype Inaba.24 It appears that as an alternate to Ogawa serotype, Inaba has appeared to aid the persistence of cholera and thus perpetuate the spread of V. cholerae El Tor.
The susceptibility patterns of V. cholerae isolates showed alarmingly high resistance (97%) towards tetracycline, whose mode of action is reversibly binding to receptors on the 30S ribosomal subunit of bacteria, preventing attachment of aminoacyl-transferable ribonucleic acid (tRNA) to the transferable ribonucleic acid (RNA)-ribosome complex, and this prevents the addition of amino acids to elongating peptide chain, preventing protein synthesis. These findings do not concur with the findings of a previous study that were reported during another outbreak in 2014 in Kisumu county.24 These findings reported that 100% of isolates were susceptible to tetracycline, a fact that should be of major concern because there was a gap of hardly 5 years between the two outbreaks. Most probably, this is attributed to the rise of antimicrobial resistance globally, as bacteria now could interchange resistant genes amongst one another because of integron genes which are described as vehicles for the acquisition of antibiotic-resistant genes.25 These findings suggest that tetracycline should not be used for the treatment of cholera infections caused by current V. cholerae O1 strains. It is thus important to monitor tetracycline-resistant patterns against V. cholerae strains, more especially at the study site, because this has been the drug of choice during cholera outbreaks,26 and therefore its preferred use could be responsible for reoccurrence of cholera in the region as fewer susceptibility tests were conducted before its administration. This finding concurs with a study performed in Mozambique on antimicrobial resistance of V. cholerae O1 isolates in 2007; the study reported high incidence of resistance to tetracycline (97.3%), used as the first-line drug for cholera treatment.27 Doxycycline, on the other hand, has been used extensively in this county for many years for cholera management. However, another study conducted in Kenya in 2007 to determine the antimicrobial response of V. cholerae isolates reported that all isolates were susceptible to doxycycline.28 In the present study, there was less resistance to doxycycline (i.e. only in 1.00% of isolates), suggesting that the antibiotic was effective in managing cholera in Kisumu County; however, close attention should be given to the emergence of these resistant strains.
A high incidence of resistance to cotrimoxazole (98.02%) was also observed in the present study, which is something to worry about. This finding is in agreement with previous studies conducted in Kenya16 and Mozambique27,29 that reported 97% and 99% resistance, respectively. Cotrimoxazole has been the first-line treatment for diarrhoea infections in Kenya, and it is commonly prescribed not only for gastrointestinal tract infections, including diarrhoea, but also for the treatment of respiratory tract infections, urinary tract infections and skin infections. Its mode of action is by targeting a subunit of DNA gyrase, which is essential in the production of bacteria DNA. It is often prescribed to immunocompromised patients, including human immunodeficiency virus (HIV) patients, which are common in the study area with a prevalence of 16.3%.13 Such a vast usage of cotrimoxazole, and the fact that it is relatively cheap and could be acquired over the counter even without a prescription, may have contributed to the emergence of resistance towards this antibiotic as observed in this study.
A high incidence of chloramphenicol-resistant isolates (44.55%) was also observed. Chloramphenicol is a bacteriostatic and broad-spectrum antibiotic effective against a wide variety of Gram-positive and Gram-negative bacteria. Its mode of action is by irreversibly binding to a receptor site on the 50S subunit of bacterial ribosome, inhibiting peptidyl transferase, consequently resulting in the prevention of amino acid transfer to growing peptide chains, leading to inhibition of protein synthesis. Up to the late 1990s, it was used as the first-line antibiotic treatment for typhoid and other Salmonella infections.30 However, because of resistance and safety issues, it is no longer the first-line treatment in enteritis. In low-income countries, it is still widely used, as it is not expensive and readily available.29 Chloramphenicol has been recommended by the World Health Organization (WHO) for the treatment of cholera in children and pregnant women.31 The high resistance observed may be explained by its frequent usage for the treatment of severe diarrhoea and other infectious diseases. Other reports in Kenya have indicated that V. cholerae O1 isolates are resistant to chloramphenicol.16. A similar incidence was observed for other pathogens causing diarrhoea in a study conducted in the study area.31
A higher resistance rate to ampicillin (89.11%) was observed compared to that towards amoxicillin (64.36%), whose mode of activity is by targeting penicillin-binding proteins - a group of enzymes found anchored in the cell membrane, involved in the cross-linking of bacterial cell wall. Cholera isolates from previous outbreaks in Kenya were known to exhibit resistance to ampicillin,13 doxycycline and streptomycin.32 There was an emergence of isolates resistant to nalidixic acid as well, with 81% resistant to this antimicrobial which has been primarily associated with the presence of IncC conjugative plasmids.30 All isolates were sensitive towards ciprofloxacin, gentamicin and ofloxacin, thus confirming the higher efficiency of these agents against V. cholerae isolates at Kisumu County.
These findings therefore clearly demonstrate the need for susceptibility tests to be carried out as some resistance cases of V. cholerae to common antibiotics used to manage enteritis and diarrhoea have been documented. The findings also demonstrate that it is high time that the usage of tetracycline and cotrimoxazole needs to be monitored as their high resistance levels are of great concern.
Acknowledgements
The first author (S.O.A) acknowledges Drs E.O. Omwenga and I.I. Daud for guiding him in the writing of this research article. Furthermore, he wants to thank Drs K.S. Musyoki and A. Munyau for their input that made this manuscript take its final shape.
Competing interests
The authors' declare that no competing interests exist.
Authors' contributions
All authors contributed equally to this work.
Funding information
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
Disclaimer
The views and opinions expressed in this article are those of the authors and do not necessarily reflect the official policy or position of any affiliated agency of the authors.
References
1.Jena PP, Gur R, Sharma A, Duggal SD, Bharara T. Antibiotic susceptibility profile of 'Vibrio Cholerae O1 Biotype El Tor Serotype Ogawa' outbreak in North Delhi, India. J Pediatr Infect Dis. 2018;13(04):277-282. https://doi.org/10.1055/s-0038-1661389 [ Links ]
2.Cairncross S, Feachem R. Environmental health engineering in the tropics: Water, sanitation and disease control. London: Routledge; 2018. [ Links ]
3.Okaka FO, Odhiambo B. Relationship between flooding and outbreak of infectious diseases in Kenya: A review of the literature. J Environ Publ Health. 2018;2018:8. https://doi.org/10.1155/2018/5452938 [ Links ]
4.Hinson R, Aziato L, Adeola O, et al. Health Service Marketing Management in Africa. Productivity Press; 2019. [ Links ]
5.Narra R, Maeda JM, Temba H, et al. Notes from the field: Ongoing cholera epidemic - Tanzania, 2015-2016. MMWR. 2017;66(6):177. https://doi.org/10.15585/mmwr.mm6606a5 [ Links ]
6.Saidi SM. Dynamics of cholera outbreaks in endemic areas of Kenya. Doctoral dissertation. 2014, Nairobi University, Kenya. [ Links ]
7.Craig RK. Cholera and climate change: Pursuing public health adaptation strategies in the face of scientific debate. Hous J Health Law Pol. 2018;18:29. [ Links ]
8.Shikanga OT, Mutonga D, Abade M, et al. High mortality in a cholera outbreak in western Kenya after post-election violence in 2008. Am J Trop Med Hyg. 2009;81(6):1085-1090. https://doi.org/10.4269/ajtmh.2009.09-0400 [ Links ]
9.Mugoya I, Kariuki S, Galgalo T, et al. Rapid spread of Vibrio cholerae O1 throughout Kenya, 2005. Am J Trop Med Hyg. 2008;78(3):527-533. https://doi.org/10.4269/ajtmh.2008.78.527 [ Links ]
10.Iro MA, Sell T, Brown N, Maitland K. Rapid intravenous rehydration of children with acute gastroenteritis and dehydration: A systematic review and meta-analysis. BMC Pediatr. 2018;18(1):44. https://doi.org/10.1186/s12887-018-1006-1 [ Links ]
11.Dalsgaard A, Forslund A, Petersen A, et al. Class 1 integron-borne, multiple-antibiotic resistance encoded by a 150-kilobase conjugative plasmid in epidemic Vibrio cholerae O1 strains isolated in Guinea-Bissau. J Clin Microbiol. 2000;38(10):3774-3779. https://doi.org/10.1128/JCM.38.10.3774-3779.2000 [ Links ]
12.Kiiru JN, Saidi SM, Goddeeris BM, Wamae NC, Butaye P, Kariuki SM. Molecular characterization of Vibrio cholerae O1 strains carrying an SXT/R391-like element from cholera outbreaks in Kenya: 1994-2007. BMC Microbiol. 2009;9(1):275. https://doi.org/10.1186/1471-2180-9-275 [ Links ]
13.Pugliese N, Maimone F, Scrascia M, Materu SF, Pazzani C. SXT-related integrating conjugative element and IncC plasmids in Vibrio cholerae O1 strains in Eastern Africa. J Antimicrob Chemother. 2009;63(3):438-442. https://doi.org/10.1093/jac/dkn542 [ Links ]
14.Wands AM, Cervin J, Huang H, et al. Fucosylated molecules competitively interfere with cholera toxin binding to host cells. ACS Infect Dis. 2018;4(5):758-770. https://doi.org/10.1021/acsinfecdis.7b00085 [ Links ]
15.Baron S, Chevalier S, Lesne J. Vibrio cholerae in the environment: A simple method for reliable identification of the species. J Health Popul Nutr. 2007;25(3):312. [ Links ]
16.Lekshmi M, Ammini P, Kumar S, Varela MF. The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms. 2017;5(1):11. https://doi.org/10.3390/microorganisms5010011 [ Links ]
17.Kovacikova G, Lin W, Taylor RK, Skorupski K. The fatty acid regulator FadR influences the expression of the virulence cascade in the El Tor biotype of Vibrio cholerae by modulating the levels of ToxT via two different mechanisms. J Bacteriol. 2017;199(7):e00762-e007816. [ Links ]
18.Folgosa E, Mastrandrea S, Cappuccinelli P, et al. Molecular identification of pathogenicity genes and ERIC types in Vibrio cholerae O1 epidemic strains from Mozambique. Epidemiol Infect. 2000;127(1):17-25. https://doi.org/10.1017/S0950268801005623 [ Links ]
19.World Health Organisation (WHO). Made in Viet Nam vaccines: Efforts to develop sustainable in-country manufacturing for seasonal and pandemic influenza vaccines: Consultation held in Viet Nam, April-June 2016. Geneva: WHO; 2017. [ Links ]
20.Oyugi EO. Male partner involvement in elimination of mother to child transmission of HIV and the associated factors in Kisumu East Sub-County, Kenya (doctoral dissertation, COHES, JKUAT). Clin Immunol. 2018;192:30-39. [ Links ]
21.Diesner SC, Bergmayr C, Wang XY, et al. Characterization of Vibrio cholerae neuraminidase as an immunomodulator for novel formulation of oral allergy immunotherapy. Clin Immunol. 2018;192:30-39. https://doi.org/10.1016/j.clim.2018.03.017 [ Links ]
22.De R, Ramamurthy T, Sarkar BL, et al. Retrospective genomic analysis of Vibrio cholerae O1 El Tor strains from different places in India reveals the presence of ctxB-7 allele found in Haitian isolates. Epidemiol Infect. 2017;145(11):2212-2220. https://doi.org/10.1017/S0950268817001182 [ Links ]
23.Musyoki SM. Roles and responsibilities for post-ODF engagement: Building an enabling institutional environment for CLTS sustainability. Sustainable Sanitation for All: Experiences, challenges, and innovations. 2016; p. 167. [ Links ]
24.Iwanaga M, Toma C, Miyazato T, Insisiengmay S, Nakasone N, Ehara M. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob Agents Chemother. 2004;48(7):2364-2369. https://doi.org/10.1128/AAC.48.7.2364-2369.2004 [ Links ]
25.Wiesner M, Zaidi MB, Calva E, Fernández-Mora M, Calva JJ, Silva C. Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains. BMC Microbiol. 2009;9(1):131. [ Links ]
26.Poulin-Laprade D, Carraro N, Burrus V. The extended regulatory networks of SXT/R391 integrative and conjugative elements and Inc A/C conjugative plasmids. Front Microbiol. 2015;6:837. https://doi.org/10.3389/fmicb.2015.00837 [ Links ]
27.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45(11):2991-3000. https://doi.org/10.1128/AAC.45.11.2991-3000.2001 [ Links ]
28.Falagas ME, Grammatikos AP, Michalopoulos A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev Anti-Infect Ther. 2008;6(5):593-600. https://doi.org/10.1586/14787210.6.5.593 [ Links ]
29.Murhekar M, Dutta S, Ropa B, Dagina R, Posanai E, Rosewell A. Vibrio cholerae antimicrobial drug resistance, Papua New Guinea, 2009-2011. WPSAR. 2013;4(3):60. https://doi.org/10.5365/wpsar.2013.4.2.002 [ Links ]
30.Onyango D, Karambu S, Abade A, Amwayi S, Omolo J. High case fatality cholera outbreak in Western Kenya, August 2010. Pan Afr Med J. 2013;15(1):109. https://doi.org/10.11604/pamj.2013.15.109.2270 [ Links ]
31.Oundo JO, Kariuki S, Maghenda JK, Lowe BS. Antibiotic susceptibility and genotypes of non-typhi Salmonella isolates from children in Kilifi on the Kenya coast. Trans Roy Soc Trop Med Hyg. 2000;94(2):212-215. https://doi.org/10.1016/S0035-9203(00)90280-3 [ Links ]
32.Scrascia M, Maimone F, Mohamud KA, et al. Clonal relationship among Vibrio cholerae O1 El tor strains causing the largest cholera epidemic in Kenya in the late 1990s. J Clin Microbiol. 2006;44(9):3401-3404. https://doi.org/10.1128/JCM.00611-06 [ Links ]
 Correspondence:
Correspondence:
Silas Awuor
silasawuor@gmail.com
Received: 07 Oct. 2019
Accepted: 24 Mar. 2020
Published: 08 Dec. 2020














