Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
Water SA
versão On-line ISSN 1816-7950
versão impressa ISSN 0378-4738
Water SA vol.49 no.4 Pretoria Out. 2023
http://dx.doi.org/10.17159/wsa/2023.v49.i4.4036
RESEARCH PAPER
Occurrence of multidrug-resistant Escherichia coli and antibiotic resistance genes in a wastewater treatment plant and its associated river water in Harare, Zimbabwe
Hilary Takawira; Joshua Mbanga
Department of Applied Biology and Biochemistry, National University of Science and Technology, P.O. Box AC 939 Ascot, Bulawayo 00263, Zimbabwe
ABSTRACT
Wastewater treatment plants (WWTPs) have been identified as point sources of antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARG). Due to variations in antibiotic use and prescribing patterns in different countries, it is imperative to establish the presence of ARB and ARGs in water environments on a country-by-country basis. This study investigated the occurrence of 11 antibiotic-resistance genes (QNRB, DFR14, CTX-M, KPC, Sul1, QNRA, Sul2, ERMB, ERMA, SHV, NDM), and antibiotic-resistant Escherichia coli in a WWTP and its associated river water in Harare, Zimbabwe. 24 water samples were collected across 3 sites: upstream and downstream of the WWTP; final effluent of the WWTP. The samples were collected weekly for 8 weeks. Pure cultures of the E. coli isolates were obtained by membrane filtration (0.45 µm) and repeated streaking on Tryptone Bile X-glucuronide followed by biochemical tests (indole test; citrate test; motility, indole, and ornithine). Antibiotic resistance profiling was done for 12 antibiotics using the disc diffusion method. Total genomic DNA was extracted from the 21 water samples and the occurrence of 11 antibiotic-resistant genes investigated using conventional PCR. 86 E. coli isolates were obtained from the sampled sites: 28 from the upstream site, 26 from the WWTP effluent, and 32 from the downstream site. The results from chi-squared analysis showed a significant association (p < 0.05) between the sampling site and the percentage of antibiotic-resistant E. coli for all 12 antibiotics investigated. The percentage of E. coli isolates resistant to the tested antibiotics varied from 29% (ertapenem) to 80.2% (ciprofloxacin). 81 (94.2%) E. coli isolates were resistant to antibiotics from >3 classes. Eight (8/11, 72.7%) ARGs were detected in the WWTP effluent and river water samples. Results indicate that the investigated WWTP and associated river water are reservoirs of ARGs and antibiotic-resistant E. coli, which is a public health concern.
Keywords: antibiotic resistance, wastewater treatment, Zimbabwe, Escherichia coli, antibiotic resistance genes
INTRODUCTION
Antibiotic resistance has become a global threat to public health systems around the world, as bacteria have developed various resistance mechanisms to antibiotics (De Kraker et al., 2016). Based on data from 204 countries, antibiotic resistance is currently estimated to be responsible for 1.27 million fatalities per year (Murray et al., 2022). Antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) have been detected and quantified from different environmentally relevant matrices, including WWTPs which are now considered hotspots for the development and dissemination of ARGs (Adefisoye and Okoh, 2016).
Wastewater reclamation facilities serve as reservoirs for antibiotic-resistance genes associated with human and animal pathogens and conventional wastewater treatment methods are unable to remove antibiotics from final effluent (Adefisoye and Okoh, 2016; Hirsch et al., 1999; Rather et al., 2017). WWTPs and surface water are repositories of multidrug-resistant E. coli isolates that harbour extended-spectrum p-lactamase (ESBL) genes (Nzima et al., 2020). In Zimbabwe, sewage and water treatment plants are not well developed and there is sometimes no final disinfection step in wastewater treatment (Nhapi et al., 2004). The catchment of the WWTP investigated in this study mainly comprises the watershed of the Mukuvisi River to the south and south east of downtown Harare. After biological nutrient removal, the wastewater from the WWTP is discharged into the river without any tertiary treatment.
Escherichia coli is a commensal bacterium that can be easily transmitted to the environment through manure, animal faeces, improperly treated wastewater or sewage, and sewage overflow caused by heavy rains (Mascher et al., 2017). E. coli can exist as a commensal in the intestinal flora of humans and animals or as a pathogen causing a range of infections. Most E. coli strains are excreted into the environment and many of them can produce ESBL enzymes (Kraemer et al., 2019; Mascher et al., 2017; Nzima et al., 2020). E. coli is readily found in water environments as a contaminant, and has thus been routinely used to study antibiotic resistance. E. coli is an excellent surrogate for investigating the spread of antibiotic resistance as it can easily acquire and transmit ARGs (Poirel et al., 2018).
Long-term monitoring of ARB in areas with treated or untreated sewage may provide an indication of the levels and development of local clinical resistance and reveal the ARB circulating within a given population (Kwak et al., 2015). However, information on the occurrence and prevalence of antibiotic-resistant E. coli and associated ARGs in wastewater effluent from WWTPs and associated river water in Zimbabwe is limited. This study, therefore, aimed to detect and characterise antibiotic-resistant E. coli and ARG genes in a wastewater treatment plant and its associated river water in Harare, Zimbabwe.
MATERIALS AND METHODS
Study site and sample collection
The WWTP catchment mainly comprises the watershed of the Mukuvisi River to the south and south east of downtown Harare. The solids (sludge) and liquid (effluent) portions of the incoming raw sewage are separated in the settling tank. The effluent overflows a circular weir and gravitates to the biological nutrient removal (BNR) unit. There is no chlorination done as the final treatment but rather the final effluent is discharged into Mukuvisi River after BNR treatment.
Sampling was done once a week from August to September 2021; thus a total of 8 samples were collected per site over a 2-month period. For each sampling event, 3 composite samples were collected upstream (Site 1), WWTP final effluent (Site 2), and downstream (Site 3) as shown in Fig. 1. Close to the downstream sampling site, there is an informal settlement. Field observations during sampling showed that the water at the downstream sampling site is used directly by the inhabitants of the informal settlement for several household purposes, including brushing teeth, bathing and washing, without any prior treatment. A total of 24 samples were collected, thus 8 samples per site over 2 months. All samples were collected using a Buerkle long handle sampler telescoop (QTE technologies, Vietnam) and aseptically transferred into 1 000 mL sterile plastic bottles (Microspec. UK/Bromborough). The samples were transported to the Water Quality Assurance Laboratory on ice to maintain a cool chain for immediate processing.
Isolation of E. coli
Ten-fold serial dilutions were done for all the samples and 100 mL was filtered. Membrane filtration was done by filtering 100 mL of each sample dilution through a 0.45 um membrane filter using a membrane filtration unit. The filter papers were placed on Petri dishes containing TBX media (Oxoid, England/Hampshire). The Petri dishes were then incubated at 37°C for 24 h. Each dilution was replicated twice. Given the possibility that a large proportion of samples may contain many individual and often unrelated isolates, a random subset of 5 individual presumptive E. coli isolates was selected from each sample. These presumptive E. coli isolates were later confirmed using biochemical tests, including the indole test, citrate test and motility, indole, and decarboxylase test.
Molecular confirmation of E. coli
DNA extraction was done using a standard heat lysis protocol (Mbanga et al., 2018). E. coli was confirmed using conventional PCR of the uidA ((3-D glucuronidase) gene. The PCR reaction consisted of a total reaction mixture of 10 µL with 5 uL of 2x Dreamtaq master mix (New England Biolabs, UK), 0.16 uL (0.4 uM) forward primer, 0.16 uL (0.4 uM) reverse primer (Inqaba Biotech, Pretoria, SA), and 2.68 uL nuclease-free water (New England Biolabs, UK), and 2 uL of DNA template was prepared and run on a MiniAmp Plus Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific, USA). The negative control comprised nuclease-free water in place of template DNA. The PCR profile was as follows: initial denaturation at 95°C for 3 min, 30 cycles (denaturation at 95°C for 30 s, annealing temperatures at 50°C for 30 s, extension at 72°C for 1 min) and then the final extension at 72°C for 5 min. E. coli ATCC 25922 was used as a positive control.
Antibiotic susceptibility testing
Twelve commercial antibiotic discs (Mast Diagnostics, Merseyside, UK) were employed for the antibiotic susceptibility testing. This was done on a bacterial suspension with the same turbidity as the 0.5 MacFarland standard, which was used for the antibiotic susceptibility testing on Mueller Hinton agar. The following antibiotics were used: cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 ug), cefepime (CPM, 30 ug), ceftriaxone (CRO, 30 ug), azithromycin (ATH, 15 µg), ampicillin (AP, 10 ug), amoxicillin/ clavulanic acid (AUG, 20 µg/10 µg), ertapenem (ETP, 10 ug), trimethoprim/sulfamethoxazole (TS, 1.25 ug/23.75 ug), tetracycline (TET, 30 ug), nalidixic acid (NA, 30 ug), and ciprofloxacin (CIP, 5 µg). The antibiotic susceptibility testing and interpretation of results was determined using Clinical and Laboratory Standards Institute (CLSI) guidelines and interpretative charts (CLSI, 2020). E. coli ATCC 25922 was used for quality control. Multi-drug resistance (MDR) was defined as non-susceptibility to 1 or more antibiotics in 3 or more drug classes.
Extraction of total genomic DNA
Total genomic DNA was extracted from the 21 wastewater samples (7 from WWTP effluent, 7 from the upstream site, and 7 from the downstream site) using the Qiagen Blood and Tissue Kit (Qiagen, Manchester, UK) according to the manufacturer's protocol with few modifications. Samples collected in Week 1 were not available and hence were excluded. The samples were first centrifuged to get a pellet, and more of the sample was added at the beginning of each subsequent centrifuging round. This was done 5 times for each sample at 12 000 r/min for 5 min. DNA was then extracted from the sample following the manufacturer's instructions. The quality of the extracted DNA was checked using agarose gel electrophoresis.
Detection of antibiotic-resistance genes in the wastewater samples
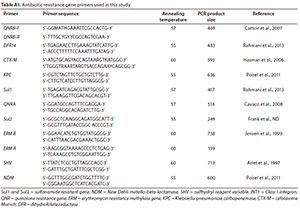
After DNA was successfully isolated directly from the 21 samples, the occurrence of antibiotic-resistance genes was investigated using conventional PCR with specific oligonucleotide primers. The ARG primers used in this study are shown in Table A1 (Appendix). A total of 11 ARGs cutting across different classes of antibiotics (p-lactams, quinolones, macrolides, trimethoprim, and sulfonamides) were assayed.
Statistical analysis
The generated data were analysed in Microsoft Excel 2018 using STATA version 17 (Stata, 2017), using tab command (to create contingency tables) to check the prevalence of resistant isolates across the three sampling sites. The prevalence of resistance for all the isolates against a panel of 12 antibiotics was recorded from the STATA outputs. To check if there was an association between the sampling site and the prevalence of resistant isolates, the chi2 command was used in STATA version 17 and this was tested at a 5% significance level, testing the null hypothesis that there was no association between the sampling site and prevalence of multidrug-resistant E. coli.
RESULTS
Isolation and identification of E. coli
A total of 86 E. coli isolates were confirmed using biochemical and molecular techniques from the 24 wastewater samples. In total, 28 isolates were from the upstream site, 26 from the final effluent of the WWTP, and 32 from the downstream site.
Prevalence of antibiotic-resistant E. coli
The antibiotic susceptibility of the tested E. coli isolates across all sampled sites is shown in Fig. 2. The highest resistance was observed towards TS (80%) and CIP (80.2%), followed by TET (79%). E. coli isolates were mostly susceptible to the carbapenem ETP (29%) (Fig 2). Resistance to third-generation cephalosporins was found to be 37.2%, 39.5%, 40.6%, and 41.8% for the antibiotics cefepime, ceftazidime, cefotaxime, and ceftriaxone, respectively. Macrolide resistance (azithromycin) was also observed in 63.9% of the E. coli isolates. A higher prevalence of antibiotic resistance was recorded in E. coli isolates from the samples that were collected downstream for all the antibiotics (Table 1). The prevalence of antibiotic-resistant isolates from the upstream samples ranged from 21% (ceftriaxone) to 57%, which was jointly recorded for tetracycline and augmentin (AUG). For the final effluent isolates, the highest (96%) frequency of antibiotic-resistant isolates was recorded for CIP, and the lowest was observed for the antibiotic ETP (32%). However, for the downstream isolates, a relatively higher prevalence of antibiotic-resistant isolates was observed compared to other sites, with the highest prevalence (97%) being recorded for the antibiotics CIP, NA, TET, AUG, and TS (Table 1). The results from chi-squared analysis showed a significant association (p < 0.05) between the sampling site and antibiotic resistance for all 12 antibiotics at a 5% significance level. Therefore, the sampling site had an effect on the prevalence of antibiotic resistance among the isolates, with the downstream site recording the highest occurrence of antibiotic resistance for 11 of the 12 antibiotics that were tested (Table 1).
Prevalence of multidrug-resistant profiles
Out of the 86 isolates that were investigated for multidrug resistance, 81 (94.2%) were resistant to 3 or more antibiotics from different classes. Only 6 isolates from the upstream site were resistant to 2 or fewer antibiotics. However, all isolates were resistant to at least one antibiotic. A total of 48 multidrug-resistance patterns were observed from the 81 MDR E. coli isolates. The most prevalent MDR patterns were AUG-TET-ATH-CRO-CPM-CTX-CAZ-CIP-NA-AP-TS and AUG-TET-ATH-CIP-NA-AP-TS that were observed for 10 isolates each (Table 2). An isolate from the downstream site was resistant to all 12 antibiotics (Table 2).
Detection of antibiotic-resistance genes
Antibiotic-resistance genes were detected directly from 21 wastewater samples. A total of 8 (72.7%) different ARGs were detected across the sampled sites (Table 3). The distribution of the ARGs ranged between 0 and 81%. Most samples were found to harbour antibiotic-resistance genes (Table 3) for 2 or more classes of antibiotics. The sul2 gene (81%) was the most prevalent ARG occurring at all 3 sampled sites. The ermA, ermB, qnrA, qnrB, and CTX-M genes also occurred at all 3 sites but at lower frequencies. The NDM gene was only detected in the final effluent of the WWTP (Table 3). The KPC, SHV, and Dfr14 genes were not detected in any of the assayed water samples.
DISCUSSION
Prevalence of antibiotic-resistant E. coli across the three sampling sites
Of the 86 isolates that were tested for antibiotic resistance, all were resistant to at least one of the 12 antibiotics tested. The isolates showed elevated resistance (>72%) to AUG, AP, NA, TET, TS, and CIP (Fig. 2). The findings are similar to those of previous studies from around the world, including South Africa (Mbanga et al., 2021), Portugal (Bessa et al., 2014), and Vietnam (Lien et al., 2017). The study by Mbanga et al. (2021) investigated the antibiotic resistance of E. coli and Enterococcus isolates from a WWTP and its associated river upstream and downstream of the WWTP. A total of 580 E. coli isolates were assayed in the study and the highest resistance was observed against AP (63.4%), TS (57.2%), and AUG (53.1%). These percentage resistances were lower than those observed in this study for AP (75.6%), TS (80%), and AUG (72%), probably due to the differences in isolate numbers used and the source of the isolates. In this study, the highest susceptibility of E. coli isolates was towards ETP (29%) (Fig. 2). Carbapenems are reserved for treating infections where quinolones and cephalosporins are not effective. The findings in this study are consistent with other studies that have reported higher susceptibility to carbapenems. Lien et al. (2017) reported very low resistance to carbapenems, with only one isolate (out of 265 E. coli) from the rural hospital effluent found to be resistant to imipenem. Similarly, a study from Poland (Kotlarska et al., 2015) on 774 E. coli isolates from raw and treated effluents of 2 WWTPs and their receiving waters revealed that all isolates were susceptible to both meropenem and imipenem. The results from this study revealed an increased rate of resistance to CTX (40.6%), NA (78.9%) and TET (79%) (Fig. 2). In contrast, a study by Adefisoye and Okoh (2016) on the prevalence of antibiotic-resistant pathogenic Escherichia coli from treated wastewater effluents in the Eastern Cape revealed a lower rate of CTX prevalence (4.5%) and reduced resistance to NA (31.4%), and TET (60.1%). However, the South African study only focused on samples from the final effluent, which might explain the observed differences. Another finding from this study was moderate resistance of E. coli to cefepime (37.2%), a fourth-generation cephalosporin. Across all three sampling sites, the resistance to cefepime ranged from 29% (upstream site) to 41% (downstream site) (Table 1). These findings are consistent with Tissera et al. (2017) who carried out a study on the isolation of ESBL producing bacteria from urban surface waters in Malaysia and found an increasing emergence of fourth-generation cephalosporins. The emergence of such species in the environment presents a public health risk, especially where communities are known to use the water sources for domestic and recreational purposes, as in the case of Mukuvisi River, especially downstream of the investigated WWTP. The presence of E. coli resistant to cephalosporins, quinolones, and carbapenems in the wastewater and associated river water observed in the current study is a cause for concern.
The high level of resistance to the fluoroquinolones nalidixic acid (78.9%) and ciprofloxacin (80.2%) reported in our study is also of concern. The sharp increase in fluoroquinolone-resistant isolates from the upstream site to the effluent and downstream sites needs further investigation.
Fluoroquinolones are considered an effective treatment option for urinary tract infections caused by E. coli. The widespread use of quinolones as powerful broad-spectrum antibiotics to treat infections in the respiratory, urinary, and digestive systems leads to their detection in WWTPs regularly around the world. Macrolides and quinolones are also among the antibiotics most consumed worldwide (Van Boeckel et al., 2014).
For most antibiotics that were tested, the results showed a higher percentage of antibiotic-resistant isolates in the final effluent and downstream samples when compared to the upstream site (Table 1). This may imply that treated effluents affect the relative abundance of antibiotic-resistant bacteria in associated surface waters, especially in cases where a final disinfection step is not included in the wastewater treatment. This is supported by the findings of Szczepanowski et al. (2009), who reported that although the wastewater treatment process reduces microbial load by over 99%, antibiotic-resistant bacteria still find their way into the environment, specifically the associated surface waters. The increase in antibiotic-resistant isolates agrees with the findings of Koczura et al. (2014) and Osinska et al. (2017), in which a higher frequency of MDR isolates was reported in the influent and downstream samples than in the upstream samples. This could be explained by the existence of informal settlements at the downstream site in our study, pointing to the possible introduction of ARB from anthropogenic activities at this site.
A total of 81 (94.2%) E. coli isolates were MDR, with a total
of 48 MDR patterns. The MDR patterns showed that there is potentially a large diversity of resistance determinants haboured by environmental E. coli. Some of the observed MDR patterns are shown in Table 2. None of the MDR patterns were common to all 3 sampled sites, with only one pattern being common to the upstream and downstream sites i.e AUG-TET-ATH-CRO-CPM-CTX-CAZ-CIP-NA-AP-TS (Table 2). Three patterns were common to isolates from the effluent and downstream sites. This points to the mobility of isolates and their clones down the river-WWTP continuum, or the constant transfer of mobile genetic elements that confer resistance to the same antibiotics. However, most of the E. coli isolates had unique MDR patterns suggesting that the sampled sites could have distinct microenvironments.
Detection of ARGs
Eight out of the eleven antibiotic resistance genes analysed were detected in the wastewater and associated river water samples (Table 3). The results showed that sulfonamide resistance was common across all 3 sampling sites. Sulfonamide resistance in Gram-negative bacteria can probably be attributed to Sull and Sul2 genes, which are carried by plasmids. In the present study, the sul2 gene was the common one, as it was observed in 17 (81%) of the samples. This is in agreement with the findings of Stange et al. (2019), who investigated the distribution of clinically relevant antibiotic-resistance genes in Lake Tai, China, and concluded that sulfonamide-resistance genes were the most common ARGs in the environment. Another study by Yan et al. (2018) investigated the occurrence of tetracyclines, sulfonamides, and quinolones and their corresponding resistance genes in the Three Gorges Reservoir in China. Their results demonstrated that sulfonamide-resistance genes were the most ubiquitous ARGs in the environment. Lye et al. (2019) carried out a study to investigate anthropogenic impacts on sulfonamide residues and sulfonamide-resistant bacteria and genes in the Larut and Sangga Besar Rivers, and reported that the sul2 gene had the highest abundance.
The qnrA and qnrB genes were also detected in this study from all sampled sites (Table 3). They are known to confer low-level resistance to fluoroquinolones in Enterobacteriaceae and they may be found on the same resistance plasmids as ESBLs (Salah et al., 2019). Research by Colomer-Lluch et al. (2014) revealed the qnrA gene as the most prevalent quinolone resistance gene detected in urban wastewater. The presence of qnrA and qnrB genes in the assayed environments agreed with the high phenotypic resistance to ciprofloxacin and nalidixic acid observed in this study.
ERMB genes detected in this study encode for ribosomal methylase, which is responsible for macrolide resistance (Enne et al., 2001). A study by Párnánen et al. (2019) investigated the prevalence of ARGs in 12 WWTPs in European countries and concluded that emrA was the most common macrolide-resistant gene detected in WWTPs. This is similar to the results obtained in this study. The incidence of the ermA and ermB genes was 52.4% for both genes. This is in agreement with the research of Wang et al. (2020), who investigated the occurrence and fate of antibiotics, antibiotic-resistance genes, and antibiotic-resistant bacteria in municipal wastewater treatment plants. The gene for macrolide resistance, emrB, was found to be prevalent in the wastewater samples in their study.
Genes that code for p-lactam resistance were the least detected in this current study, with only CTX-M being detected in only 10 (47.6%) of the samples (Table 3). Although the p-lactam class, involving penicillins and cephalosporins which are the top two most-consumed antibiotics worldwide (Van Boeckel et al., 2014), these compounds are not frequently detected in WWTPs because members of p-lactam antibiotics readily hydrolyze owing to unstable p-lactam rings. The presence of antibiotics in WWTPs contributes to the development of antimicrobial resistance. However, research has shown that the ARGs commonly detected in WWTPs include the genes resistant to p-lactam such as blaCTX-M, blaTEM, and blaSHV.
NDM-1 is an enzyme that makes bacteria resistant to a broad range of beta-lactam antibiotics including antibiotics of the carbapenems which are a mainstay for the treatment of antibiotic-resistant bacterial infections (Van Boeckel et al., 2014). It encodes p-lactamase enzymes called carbapenemases. The most common bacteria that make these enzymes are Gram-negative K. pneumoniae and E. coli, but this can be spread from one strain of bacteria to another through horizontal gene transfer (Van Boeckel et al., 2014). However, it's unclear whether and how much of the existing ARGs are carried intercellularly (iARGs) by living, metabolically active bacteria. To go beyond qualitative and quantitative ARG analyses and gain a more comprehensive understanding of ARG dynamics, it is vital to determine which of the discovered ARGs are expressed.
CTX-M genes were detected in lower frequencies across the sites. This is in contrast with a study done in South Africa (Adegoke et al., 2020), where blaCTX-M was found in 52.6% (n = 38) of the samples. NDM genes have also been reported in the current study but at very low frequencies across the sampling sites. A study done in China also confirmed the occurrence of blaNDM-5 in wastewater (Zhang et al., 2016).
CONCLUSION
The results of our study indicate that a high proportion of MDR E. coli were present in the investigated WWTP and its receiving waters. The E. coli had a great diversity of antibiotic-resistance profiles, implying that the WWTP and its receiving river are important reservoirs of ARGs and antibiotic-resistant bacteria, which are potential human and animal pathogens. Antibiotic-resistance genes belonging to several drug classes were detected at the sampled sites directly from wastewater. The presence of antibiotic-resistant E. coli and ARGs in environmental waters presents a significant health risk to the people and animals that use these water sources and could be potential reservoirs for the dissemination of antibiotic resistance.
AUTHOR CONTRIBUTIONS
Co-conceptualized the study: JM, HT. Performed the experiments: HT, JM. Analysed the data: HT, JM. Wrote the paper: JM, HT. Supervision: JM. Undertook critical revision of the manuscript: All.
ACKNOWLEDGEMENTS
Special thanks are extended to Miss Gladys Manyembere (Microbiologist: City of Harare).
FUNDING
There was no specific funding for this work.
COMPETING INTERESTS
The authors declare that they have no conflicting interests regarding the publication of this paper.
ETHICAL APPROVAL
Ethical approval was received from the NUST Institution Research Board (NUST/IRB/202/34) of the National University of Science and Technology (NUST).
ORCID
Joshua Mbanga: https://orcid.org/0000-0001-6592-2338
REFERENCES
ADEFISOYE MA and OKOH AI (2016) Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiol. Open5 143-151. https://doi.org/10.1002/mbo3.319 [ Links ]
ADEGOKE AA, MADU CE, AIYEGORO OA, STENSTRÖM TA and OKOH AI (2020) Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Res. Infect. Control. 9. https://doi.org/10.1186/s13756-020-0702-4 [ Links ]
BESSA LJ, BARBOSA-VASCONCELOS A, MENDES Â, VAZ-PIRES P and DA COSTA PM (2014) High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J. Water Health 12 426-435. https://doi.org/10.2166/wh.2014.160 [ Links ]
COLOMER-LLUCH M, JOFRE J and MUNIESA M (2014) Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J. Antimicrob. Chemother. 69 1265-1274. https://doi.org/10.1093/jac/dkt528 [ Links ]
CLSI. CLSI M100-ED30:2020 Performance Standards for Antimicrobial Susceptibility Testing, 30th Edition [Internet]. Wayne, PA; 2020. URL: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSIM100ED30:2020 (Accessed 30 July 2021). [ Links ]
DE KRAKER ME, STEWARDSON AJ and HARBARTH S (2016) Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 13. https://doi.org/10.1371/journal.pmed.1002184 [ Links ]
ENNE VI, LIVERMORE DM, STEPHENS P and HALL LMC (2001) Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet. 357 1325-1328. https://doi.org/10.1016/S0140-6736(00)04519-0 [ Links ]
HIRSCH R, TERNES T, HABERER K and KRATZ K-L (1999) Occurrence of antibiotics in the aquatic environment. Sci. Total Environ. 225 109-118. https://doi.org/10.1016/s0048-9697(98)00337-4 [ Links ]
KOCZURA R, PRZYSZLAKOWSKA B, MOKRACKA J and KAZNOWSKI A (2014) Class 1 integrons and antibiotic resistance of clinical Acinetobacter calcoaceticus-baumannii complex in Poznan, Poland. Curr. Microbiol. 69 258-262. https://doi.org/10.1007/s00284-014-0581-0 [ Links ]
KOTLARSKA E, ANETA L, PISOWACKA M and BURZYNSKI A (2015) Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 22 2018-2030. https://doi.org/10.1007/s11356-014-3474-7 [ Links ]
KRAEMER SA, RAMACHANDRAN A and PERRON GG (2019) Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 7. https://doi.org/10.3390/microorganisms7060180 [ Links ]
KWAK YK, COLQUE P, BYFORS S, GISKE CG, MÖLLBY R and KÜHN I (2015) Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents 45 25-32. https://doi.org/10.1016/JlJANTIMICAG.2014.09.016 [ Links ]
LIEBANA E, CARATTOLI A, COQUE TM, HASMAN H, MAGIORAKOS A-P, MEVIUS D, PEIXE L, POIREL L, SCHUEPBACH-REGULA G, TORNEKE K and co-authors (2013) Public health risks of enterobacterial isolates producing extended-spectrum p-lactamases or AmpC β-lactamases in food and food-producing animals: An EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin. Infect. Dis. 56 1030-1037. https://doi.org/10.1093/cid/cis1043 [ Links ]
LIEN LTQ, LAN PT, CHUC NTK, HOA NQ, NHUNG PH, THOA NTM, DIWAN V, TAMHANKAR AJ and LUNDBORG CS (2017) Antibiotic resistance and antibiotic resistance genes in Escherichia coli isolates from hospital wastewater in Vietnam. Int. J. Env. Res. Public Health 14. https://doi.org/10.3390/ijerph14070699 [ Links ]
LYE YL, BONG CW, LEE CW, ZHANG RJ, ZHANG G, SUZUKI S and CHAI LC (2019) Anthropogenic impacts on sulfonamide residues and sulfonamide resistant bacteria and genes in Larut and Sangga Besar River, Perak. Sci. Total Environ. 688 1335-1347. https://doi.org/10.1016/j.scitotenv.2019.06.304 [ Links ]
MASCHER F, MASCHER W, PICHLER-SEMMELROCK F, REINTHALER FF, ZARFEL GE and KITTINGER C (2017) Impact of combined sewer overflow on wastewater treatment and microbiological quality of rivers for recreation. Water 9. https://doi.org/10.3390/w9110906 [ Links ]
MBANGA J, ABIA AL, AMOAKO D and ESSACK SY (2021) Longitudinal surveillance of antibiotic resistance in Escherichia coli and Enterococcus spp. from a wastewater treatment plant and its associated waters in KwaZulu-Natal, South Africa. Microb. Drug Resist. 27 904-918. https://doi.org/10.1089/mdr.2020.0380 [ Links ]
MBANGA J, SIBANDA A, RUBAYAH S, BUWERIMWE F and MAMBODZA K (2018) Multi-drug resistant (MDR) bacterial isolates on close contact surfaces and health care workers in intensive care units of a tertiary hospital in Bulawayo, Zimbabwe. J. Adv. Med. Med. Res. 27 1-15. https://doi.org/10.9734/JAMMR/2018/42764 [ Links ]
MURRAY CJ, IKUTA KS, SHARARA F, SWETSCHINSKI L, ROBLES AGUILAR G, GRAY A, HAN A, BISIGNANO C, RAO C, WOOL P and co-authors (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 629-655. https://doi.org/10.1016/S0140-6736(21)02724-0. [ Links ]
NHAPI I, SIEBEL MA and GIJZEN HJ (2004) The impact of urbanisation on the water quality of lake chivero, Zimbabwe. Water Environ. J. 18 44-49. https://doi.org/10.1111/j.1747-6593.2004.tb00492.x [ Links ]
NZIMA B, ADEGOKE AA, OFON UA, AL-DAHMOSHI HOM, SAKI M, NDUBUISI-NNAJI UU and INYANG CU (2020) Resistotyping and extended-spectrum beta-lactamase genes among Escherichia coli from wastewater treatment plants and recipient surface water for reuse in South Africa. New Microbes New Infect. 38. https://doi.org/10.1016/j.nmni.2020.100803 [ Links ]
ORT C, VAN NUIJS ALN, BERSET JD, BIJLSMA L, CASTIGLIONI S, COVACI A, DE VOOGT P, EMKE E, FATTA-KASSINOS D, GRIFFITHS P and co-authors (2014) Spatial differences and temporal changes in illicit drug use in Europe quantified by wastewater analysis. Addiction 109 1338-1352. https://doi.org/10.1111/add.12570 [ Links ]
OSINSKA A, KORZENIEWSKA E, HARNISZ M and NIESTEPSKI S (2017) The prevalence and characterization of antibiotic-resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci. Total Environ. 577 367-375. https://doi.org/10.1016/j.scitotenv.2016.10.203 [ Links ]
PANDIT R, AWAL B, SHRESTHA SS, JOSHI G, RIJAL BP and PARAJULI NP (2020) Extended-Spectrum p -Lactamase (ESBL) Genotypes among multidrug-resistant uropathogenic Escherichia coli clinical isolates from a teaching hospital of Nepal. Interdiscip. Perspect. Infect. Dis. 2020 Article ID 6525826, 8 pp. https://doi.org/10.1155/2020/6525826 [ Links ]
PÀRNÀNEN KM, NARCISO-DA-ROCHA C, KNEIS D, BERENDONK TU, CACACE D, THUY DO T, ELPERS C, FATTA-KASSINOS D, HENRIQUES I, JAEGER T, KARKMAN A and co-authors (2019) Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5. https://doi.org/10.1126/sciadv.aau9124 [ Links ]
POIREL L, MADEC J-Y, LUPO A, SCHINK A-K, KIEFFER N, NORDMANN P and SCHARZ S (2018) Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6 1-27. https://doi.org/10.1128/microbiolspec.arba-0026-2017 [ Links ]
RATHER IA, KIM BC, BAJPAI VK and PARK YH (2017) Self-medication and antibiotic resistance: Crisis, current challenges, and prevention. Saudi J. Biol. Sci. 24 808-812. https://doi.org/10.1016/j.sjbs.2017.01.004 [ Links ]
SALAH FD, SOUBEIGA ST, OUATTARA AK, SADJI AY, METUOR-DABIRE A, OBIRI-YEBOAH D, BANLA-KERE A, KAROU S and SIMPORE J (2019) Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob. Resist. Infect. Control 8. https://doi.org/10.1186/s13756-019-0552-0 [ Links ]
STANGE C, YIN D, XU T, GUO X, SCHÀFER C and TIEHM A (2019) Distribution of clinically relevant antibiotic resistance genes in Lake Tai, China. Sci. Total Environ. 655 337-346. https://doi.org/10.1016/j.scitotenv.2018.11.211 [ Links ]
STATA CORP (2017) Stata Statistical Software: Release 15. StataCorp LLC, College Station TX. [ Links ]
SZCZEPANOWSKI R, LINKE B, KRAHN I, GARTEMANN KH, GÜTZKOW T, EICHLER W, PÜHLER A and SCHLÜTER A (2009) Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155 2306-2319. https://doi.org/10.1099/mic0.028233-0 [ Links ]
TEKINER lH and ÖZPINAR H (2016) Occurrence and characteristics of extended spectrum beta-lactamases-producing enterobacteriaceae from foods of animal origin. Braz. J. Microbiol. 47 444-451. https://doi.org/10.1016/j.bjm.2015.11.034 [ Links ]
TISSERA K, LIYANAPATHIRANA V, DISSANAYAKE N, PINTO V, EKANAYAKE A, TENNAKOON M, ADASOORIYA D and NANAYAKKARA D (2017) Spread of resistant gram negatives in a Sri Lankan intensive care unit. BMC Infect. Dis. 17. https://doi.org/10.1186/s12879-017-2590-7 [ Links ]
VAN BOECKEL TP, GANDRA S, ASHOK A, CAUDRON Q, GRENFELL BT, LEVIN SA and LAXMINARAYAN R (2014) Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 14 742-750. https://doi.org/10.1016/S1473-3099(14)70780-7 [ Links ]
WANG J, CHU L, WOJNÁROVITS L and TAKÁCS E (2020) Occurrence and fate of antibiotics, antibiotic resistance genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 744. https://doi.org/10.1016/j.scitotenv.2020.140997 [ Links ]
YAN M, XU C, HUANG Y, NIE H and WANG J (2018) Tetracyclines, sulfonamides and quinolones and their corresponding resistance genes in the Three Gorges Reservoir, China. Sci. Total Environ. 632 840-848. https://doi.org/10.1016/j.scitotenv.2018.03.085 [ Links ]
ZHANG F, XIE L, WANG X, HAN L, GUO X, NI Y, QU H. and SUN J (2016) Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front. Microbiol. 7. https://doi.org/10.3389/fmicb.2016.00424 [ Links ]
 Correspondence:
Correspondence:
Joshua Mbanga
Email:joshmbanga@gmail.com; joshua.mbanga@nust.ac.zw
Received: 30 October 2022
Accepted: 6 September 2023
APPENDIX