Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Water SA
On-line version ISSN 1816-7950
Print version ISSN 0378-4738
Water SA vol.48 n.4 Pretoria Oct. 2022
http://dx.doi.org/10.17159/wsa/2022.v48.i4.3941
RESEARCH PAPER
Rapid detection of drug-resistant Escherichia coli by Vitek 2 compact system
F TshabuseI; N ButheleziII; AM FolamiII; L DonnellyIII; FM SwalahaII
IDepartment of Biochemistry and Microbiology, University of Zululand, Private Bag X1001, kwaDlangezwa 3886, South Africa
IIDepartment of Biotechnology and Food Science, Durban University of Technology, PO Box 1334, Durban 4000, South Africa
IIIUmgeni Water, PO Box 9, Pietermaritzburg 3200, KwaZulu-Natal, South Africa
ABSTRACT
Sewage treatment facilities aim to reduce biological contaminants such as pathogenic bacteria, fungi, protozoa, and viruses in wastewaters before discharging them to the receiving water bodies. However, several studies have shown the persistence of these contaminants throughout the sewage treatment process. In this study, the Vitek 2 compact system was used to detect the presence of Escherichia coli in three sewage treatment facilities located in the Pietermaritzburg urban area (South Africa), and its susceptibility to antimicrobial agents. E. coli has been recognized as an important Gram-negative rod-shaped human pathogen. The effluent and influent samples were analysed to determine the fate of E. coli and its susceptibility to 17 antimicrobial agents. The system identified the presence of drug-resistant E. coli in all of the tested samples, with the highest susceptibility being to ampicillin (33%) and trimethoprim/sulfamethoxazole (27%). The Vitek 2 compact system is a quick and powerful tool to identify antimicrobial-resistant bacteria in effluents and monitoring by this systemcan be used to prevent the outbreak of waterborne diseases.
Keywords: antibiotic resistance, bacterial identification, susceptibility, Vitek 2, wastewater treatment
INTRODUCTION
Wastewater is defined as water that consists of stormwater runoff, industrial, domestic, or commercial sewage, or any mixture thereof (Naidoo and Olaniran, 2014). This wastewater contains contaminants such as nutrients, microorganisms, and organic matter, as well as contaminants of emerging concern such as pharmaceuticals and personal care products (Ginebreda et al., 2010). Wastewater treatment processes are therefore designed to either remove or reduce these contaminants to acceptable levels before the effluent is discharged to the receiving water body (Frigon et al., 2013). However, several studies have revealed the ability of most waterborne pathogens, such as Escherichia coli, to survive the chlorination method and thus be released into the environment (Jjemba et al., 2010; Ramirez-Castillo et al., 2015), where, if ingested, inhaled or encountered by susceptible individuals, it can cause waterborne diseases (Leclerc et al., 2002).
According to the World Health Organisation (WHO, 2018), about 844 million people lack safe potable water, with 159 million people dependent on surface waters. Contaminated water is linked to different types of infectious diseases, such as gastroenteritis, cholera, salmonellosis, typhoid, hepatitis and dysentery, with diarrhoea as the common symptom (Colvin et al., 2016). It is estimated that 842 000 people die each year from diarrhoea, of which 361 000 are children younger than the age of 5 years (WHO, 2018). Globally, these diseases are estimated to cause an economic loss of 12 billion USD per annum (Alhamlan et al., 2015).
Furthermore, several studies have revealed the inability of wastewater treatment processes to remove antibiotic-resistant genes (ARG) (Shejale et al., 2020; Nguyen et al., 2021). This is a concern since the nutrient-rich activated sludge has been shown to be a perfect environment for transfer of antibiotic-resistant genes between bacteria (Berendonk et al., 2015; Di Cesare et al., 2016; Hembach et al., 2017). Thus, this can lead to the development of antimicrobial-resistant bacteria which can be discharged into the aquatic environment. Recently, antimicrobial-resistant bacteria have also been detected in treated effluents (Alexander et al., 2020); this is posing as a new concern threatening the healthcare system of the 21st century. Between the year 2014 and 2016, over 1 million people has been reported to have died due to conditions in which antibiotic-resistant bacteria were implicated (Alexander et al 2020), and this has been projected to increase in the coming decades (Humphreys and Fleck 2016). Therefore, improving water quality is a global goal that needs immediate attention, of which surveillance programmes can be a starting point.
However, such surveillance programmes are usually lacking in developing countries, including South Africa. Molecular methods which are quick and accurate are critically required in detecting and tracing E. coli in animals, food, and water, to minimize the size and number of waterborne disease outbreaks in developing countries (Fratamico et al., 2016). Bacterial detection and antimicrobial sensitivity testing by Vitek 2 compact system have been successfully conducted in positive blood cultures (Bazzi et al., 2017). In this study, Vitek 2 compact system was used to detect the presence of Escherichia coli (E. coli) in three sewage treatment facilities located in the Pietermaritzburg urban area (South Africa) and its susceptibility to antimicrobial agents. However, usage of this technique in wastewater treatment plant samples has been minimal to date and its application requires more research, since it is a robust, reliable technique offering both microbial identification and AST. The results of this study revealed that Vitek 2 compact system is a quick and powerful tool to identify antimicrobial-resistant bacteria in the wastewater treatment plant effluents, and monitoring by this method can be used to prevent the outbreak of waterborne diseases.
METHODS
Wastewater sample collection and processing
The wastewater samples were collected monthly from August to December 2020 from 3 wastewater treatment plants around Pietermaritzburg, KwaZulu-Natal. Plant A (29°36'04.9"S 30°25' 44.9"E) was the largest of the three treatment plants and serves the Msunduzi Local Municipality, whereas Plant B (29°29'36.0"S 30°14'01.6"E) serves Howick town and Plant C (29°41'01.9"S 30°27'53.4"E) services part of the Ashburton area (Fig. 1). All these wastewater treatment plants use chlorination as the chemical process of deactivating the microorganisms. Samples were collected into 500 mL sterile plastic bottles containing sodium thiosulphate to neutralize chlorine in effluents. The temperature and pH values were measured on-site using a portable pH meter (Hach, S.A). A sample volume of 100 mL of influent and 200 mL of effluent was used for the determination of bacterial concentration via the membrane filtration method (0.45 μm, Millipore, U.S) and the filtrate was then stored in phosphate buffer saline (PBS) at 4°C for downstream experimentation.
Determination of BOD and COD in wastewater samples
Standard methods (Baird et al., 2012) were used to determine the biological oxygen demand (BOD) values. Briefly, the optical density (OD) values were measured before and after the incubation period (at 21°C for 5 days), and the difference gave the BOD values. For chemical oxygen demand (COD), the wastewater samples were digested (for 2 h at 148°C) in a solution of potassium dichromate, sulphuric acid, silver sulphate and mercuric sulphate contained in the COD tubes. Following the digestion period, the samples were allowed to cool at room temperature and the COD values were read using a photometer.
Enumeration of E. coli
A sample volume of 100 mL was used to estimate the number of E. coli cells, according to the manufacturer's instructions, using the Colilert Quanti-Tray/2000 system (IDDEX, USA). Following the incubation period (22 h at 37°C), the Quanti-Trays were examined under long wave (366 nm) ultraviolet (UV) light, and wells that turned yellow and fluoresced were identified as E. coli positive and selected for AST analysis. Efficiency of WWTPs in removing E. coli and percentage and log reductions were calculated using Eqs 1 and 2, respectively (Microchemlab, 2021).
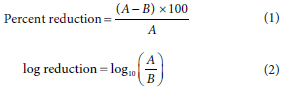
where A denotes the number of viable microorganisms before treatment, and B is the number of viable microorganisms after treatment.
Antimicrobial susceptibility test (AST) with Vitek 2 compact system
The quanti-Tray/2000 were disinfected using 70% ethanol and yellow and fluorescing (E. coli) wells were selected and aseptically punctured using sterile tips and sub-cultured onto XM-G agar (Hyserve, Germany) for 20 h at 37°C. The presumptive E. coli colonies (blue or blue purple) were transferred onto nutrient agar (Oxoid, UK) and incubated at 37°C for 24 h. Following the incubation period, the bacterial isolates were emulsified in a 3 mL 0.45% saline solution to the density of 0.5 to 0.63 McFarland measurement using DensiCHEK Plus instrument (Biomerieux, USA). The prepared suspensions were used for both bacterial identification and AST using the Vitek 2 compact system (Biomerieux, USA) according to the manufacturer's instructions (Biomerieux, USA) - the Gram negative (GN) card was used for bacterial identification, and AST-N256 card was used for AST with Vitek 2 compact system. The selected E. coli isolates were tested against 17 antibiotics representing aminoglycoside, penicillin, carbapenems, cephalosporins, quinolones, tetracyclines and sulphonamides (Table 1). The AST-N256 card was automatically filled by a vacuum device, sealed, and inserted into the Vitek 2 reader-incubator module (incubation temperature: 35.5°C), and subjected to a kinetic fluorescence measurement for identification. Turbidity was measured as an indication of susceptibility every 15 min. At the completion of the incubation cycle, bacterial isolates were identified, and MIC values were determined for each antibiotic contained on the card. The E. coli ATCC 25922 was used as a positive control and the results were interpreted following the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020).
Antimicrobial susceptibility testing using agar diffusion
The E. coli isolates were evenly spread onto the surface of nutrient agar and the disks were applied to the plate. After the incubation period (37°C for 20 h), the minimum inhibitory concentrations were recorded by determining the diameter of the inhibition zone using a ruler. The experiment was performed in duplicate.
Amplification of malate dehydrogenase (mdh) gene by colony PCR
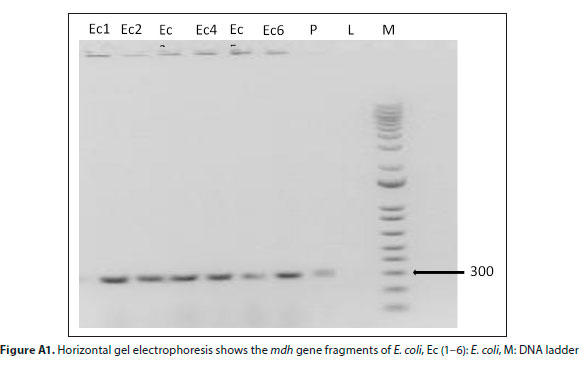
Colony PCR was performed on the bacterial isolates for the molecular confirmation of isolated E. coli in a Bioer XP thermal cycler in a total reaction volume of 25 μL. Each reaction contained 12.5 μL2x Biolabs PCR master mix (containing DNA polymerase, PCR buffer and dNTPs), 2 μL 0.5 mM of the mdh forward (GGTATGGATCGTTCCGACCT) and reverse (GGCAGAATGGTAACACCAGAGT) primers (Omar and Barnard, 2010), 2 |L template DNA and PCR grade water to a volume of 25 μL. The mdh gene was amplified following modified Tarr et al. (2002) PCR conditions: initial denaturation step at 95°C for 15 min, followed by a 30-cycle reaction (consisting of denaturing at 95°C for 45 s, annealing at 65°C for 1 min 5 s, extension at 72°C for 2 min) and final elongation at 72°C for 5 min. The PCR amplicon was resolved on ethidium bromide-stained horizontal agarose gel (1.5 %, w/v) and electrophoresis was allowed to run for 1 h in electric field strength of 100 V using 1x TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA). The gel was visualized under UV light (BioRad, USA). The relative sizes of the DNA fragments were compared with molecular 100 bp marker run with the amplicons.
RESULTS AND DISCUSSION
Physicochemical properties of wastewater samples
BOD is a universal determinant of organic quality of water, and higher values (above 5 mgO2/L) indicate poorer water quality (Lokman et al., 2020). In this study, the BOD values ranged from 85.6 to 283.0 mgO2/L for the influent and 1.4 to 11.6 for the effluent while COD concentrations (mgO2/L) ranged between 337 and 876 for the influent and 24.0 and 93.8 for the effluent over the period of 7 sampling events (Table 2). The highest pH value (8.0) was recorded in the influent of WWTP B while the temperatures (°C) ranged from 18.0 to 24.3 for the influent and 11.5 to 19.0 for the effluent. The BOD values (mgO2/L) ranged from 29.6 to 208.0 for the influent and 1.4 to 11.6 for the effluent while COD concentrations (mgO2/L) ranged between 658.9 and 1 023.0 for the influent and 20.8 and 66.8 for the effluent (Table 2). The pH values of wastewater samples at WWTP C ranged from 7.69 to 8.46 for the influent and 7.12 to 7.70 for the effluent while the temperatures (°C) ranged from 13.4 to 20.5 for the influent and 12.9 to 22.2 for the effluent. The BOD values (mgO2/L) ranged from 93.4 to 258.0 for the influent and <1.0 to 6.4 for the effluent while COD concentrations (mgO2/L) ranged between 354.0 and 791.0 for the influent and 44.3 and 97.8 for the effluent.
Log reduction of E. coli by wastewater treatment process
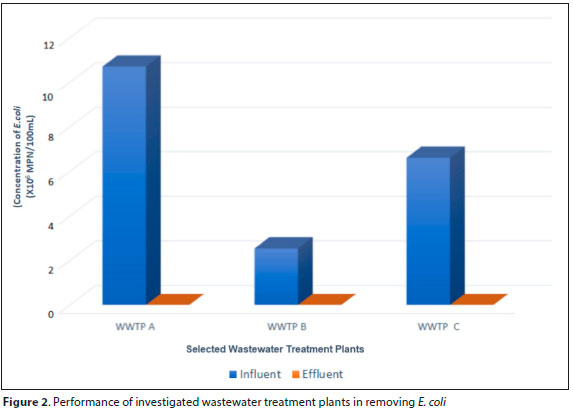
The concentration of E. coli in the influent collected from wastewater treatment process is shown in Fig. 2. WWTP A had a higher E. coli cell counts with a 4.6 log reduction, followed by WWTP B with a log reduction of 5.03 and WWTP C with a log reduction of 5.5. Taken together, it can be concluded that the three study sites had an average removal percentage of 99.999%. The results showed that the wastewater treatment processes employed within the chosen sampling sites are efficient in reducing the E. coli numbers; however, a concerning amount of E. coli cells persist throughout the treatment processes. A similar study by Brechet et al. (2014) revealed the release of a large number of E. coli in the treated effluent of Besancon city into the environment.
Identification of E. coli by Vitek 2 compact system
E. coli was positively identified in all tested samples, 59 from WWTP A, 36 from WWTP B and 40 from WWTP C using a Vitek 2 compact system. This was further confirmed by amplification of a 300 bp band fragment of mdh gene (Fig. A1, Appendix) on agarose gel, which corresponds to the presence of both commensal and pathogenic E. coli (Omar and Barnard, 2010). Furthermore, Anastasi et al. (2012) and Anastasi et al. (2013) have demonstrated the ability of pathogenic E. coli to survive the chlorination and UV stages of wastewater treatment plants. Therefore, new disinfection methods for complete inactivation of pathogenic microorganisms are urgently needed.
Antimicrobial-resistant E. coli identified by Vitek 2 compact system in wastewater
The ubiquity of antimicrobial-resistant E. coli was assessed for 17 antimicrobial agents (Table 2). As shown in Fig. 3, in WWTP A, the AM and SXT had > 20% resistance whereas the other antimicrobial agents showed between 0 and 5%. In WWTP B, the percentage resistance of AM and SXT was revealed as > 40%, whereas that of the others ranged between 0 and 15%. In WWTP C, both AM and SXT had the highest percentage resistance of > 40% and > 55%, respectively. The percentage resistance of the other antimicrobial agents ranged between 5% and 20%. It is interesting to note that AM and SXT experienced > 40% resistance in both WWTP B and WWTP C. furthermore, the data also showed WWTP C to be highly resistant to all the tested antimicrobial agents compared to WWTP A and WWTP B. Taken together, the data showed the highest prevalence of resistance in WWTP C, WWTP B and WWTP A to AM and SXT antimicrobial agents. This was also confirmed by the disk diffusion method (Fig. A2, Appendix). Ampicillin is a common antibiotic used for the treatment of E. coli infection in humans and livestock, and thus has a higher chance of selecting for resistant bacteria in the gut, including E. coli. According to the National Department of Health (2018), a quarter of antibiotics used to treat Pneumonocystis jiroveci in HIV patients in South Africa composed of trimethoprim, a constituent of co-trimoxazole (SXT). This high usage can likely induce resistance to gut microorganisms including E. coli. Contrastingly, our results also showed 100% susceptibility towards carbapenems, amikacin, and tigecycline. This was to be expected since such antibiotics are being categorised as watch-and-reserve antibiotics by the National Department of Health (2018), and thus are sparingly used.
Furthermore, the efficiency of the targeted wastewater treatment plants in the removal of antibiotic resistant E. coli was also studied. The data revealed the complete elimination of E. coli resistant to FOX, TZP and AMC (Fig. 4). However, resistance to AM and SXT was still present in the effluent. Reinthaler et al. (2003) and Rodriquez et al. (2019) have shown the contribution of sewage treatment plants in the dissemination of antibiotic-resistant microorganisms in the environment.
In conclusion, the results of this study have revealed the potential use of the Vitek 2 compact system in monitoring wastewater samples for the presence of antimicrobial resistance E. coil. This method is not only confined to the detection of E. coli but can be used in the identification of other waterborne pathogens including Salmonella and Vibrio cholerae in wastewater samples. Since water released into the aquatic environment can be used for human activities including swimming, and fishing, understanding the nature of pathogens released is important in order to prevent waterborne disease outbreaks.
ACKNOWLEDGEMENTS
The authors would like to thank the Durban University of Technology for financial assistance and Umgeni Water for their collaboration.
REFERENCES
ALEXANDER J, HEMBACH N and SCHWARTZ T (2020) Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 10 8952. https://doi.org/10.1038/s41598-020-65635-4 [ Links ]
ALHAMLAM FS, AL-QAHTANI AA and AL-AHDAL MNA (2015) Recommended advanced techniques for waterborne pathogen detection in developing countries. J. Infect. Dev. Countries. 9 128-135. https://doi.org/10.3855/jidc.6101 [ Links ]
ANASTASI EM, MATTHEWS B, STRATTON HM and KATOULI M (2012) Pathogenic Escherichia coli found in sewage treatment plants and environmental waters. Appl. Environ. Microbiol. 78 (16) 5536-5554. https://doi.org/10.1128/AEM.00657-12 [ Links ]
ANASTASI EM, WOHLSEN TD, STRATTON HM and KATOULI M (2013) Survival of Escherichia coli in two sewage treatment plants using UV irradiation and chlorination for disinfection. Water Res. 47 (17) 6670-6679. https://doi.org/10.1016/Lwatres.2013.09.008 [ Links ]
BAIRD RB, EATON AD and CLESCERI LS (2012) Standard Methods for the Examination of Water and Wastewater (Vol. 10). American Public Health Association, Washington, DC. [ Links ]
BAZZI AM, RABAAN AA, FAWARAH MM and AL-TAWFIQ JA (2017) Direct identification and susceptibility testing of positive blood cultures using high speed cold centrifugation and Vitek II system. J. Infect. Public Health. 10 (3) 299-307. https://doi.org/10.1016/j.jiph.2016.05.012 [ Links ]
BERENDONK T, MANAIA C, MERLIN C, FATTA-KASSINOS D, CYTRYN E, WALSH F, BURGMANN H, SORUM H, NORSTROM M, PONS M-N, KREUZINGER N, HUOVINEN P, STEFANI S, SCHWARTZ T, KISAND V, BAQQUERO F and MARTINEZ JL (2015) Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 310-317. https://doi.org/10.1038/nrmicro3439 [ Links ]
BRÉCHET C, PLANTIN J, SAUGET M, THOUVEREZ M, TALON D, CHOLLEY P, GUYEUX C, HOCQUET D and BERTRAND X (2014) Wastewater treatment plants release large amounts of extended-spectrum pMactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 58 (12) 1658-1665. https://doi.org/10.1093/cid/ciu190 [ Links ]
CLSI (Clinical and Laboratory Standard Institute) (2020) Performance Standards for Antimicrobial Susceptibility Testing (30th edn). CLSI supplement M100. Clinical and Laboratory Standard Institute, Wayne, Pennsylvania. [ Links ]
COLVIN C, MURUVEN D, LINDLEY D, GORDON H and SCHACHTSCHNEIDER K (2016) Water facts and futures: rethinking South Africa's water future. WWF-SA, Johannesburg. 96 pp. [ Links ]
DI CESARE A, ECKERT EM, D'URSO S, BERTONI R, GILLAN DC, WATTIEZ R and CORNO G (2016) Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 94 (1) 208-2014. https://doi.org/10.1016/j.watres.2016.02.049 [ Links ]
FRATAMICO PM, DEBROY C and NEEDLEMAN DS (2016) Emerging approaches for typing, detection, characterization, and traceback of Escherichia coli. Front. Microbiol. 7 2089. https://doi.org/10.3389/fmicb.2016.02089 [ Links ]
FRIGON D, BISWAL BK, MAZZA A, MASSON L and GEHR R (2013) Biological and physicochemical wastewater treatment processes reduce the prevalence of virulent Escherichia coli. Appl. Environ. Microbiol. 79 (3) 835-844. https://doi.org/10.1128/AEM.02789-12 [ Links ]
GINEBREDA A, MUÑOZ I, DE ALDA ML, BRIX R, LÓPEZ-DOVAL J and BARCELÓ D (2010) Environmental risk assessment of pharmaceuticals in rivers: relationships between hazard indexes and aquatic macroinvertebrate diversity indexes in the Llobregat River (NE Spain). Environ. Int. 36 (2) 153-162. https://doi.org/10.1016/j.envint.2009.10.003 [ Links ]
GLANTZ MH (2018) Water security in a changing climate. Post-Soviet Issues. 5 (3) 218-223. https://doi.org/10.24975/2313-8920-2018-5-3-218-223 [ Links ]
HEMBACH N, SCHMID F, ALEXANDER J, HILLER C, ROGALL ET and SCHAWRTZ T (2017) Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front. Microbiol. 8 (1282) 1-11. https://doi.org/10.3389/fmicb.2017.01282 [ Links ]
HUMPHREYS G and FLECK F (2016) United Nations meeting on antimicrobial resistance. Bull. World Health Organ. 94 638-639. https://doi.org/10.2471/BLT.16.020916 [ Links ]
JJEMBA PK, WEINRICH LA, CHENG W, GIRALDO E and LECHEVALLIER MW (2010) Regrowth of potential opportunistic pathogens and algae in reclaimed-water distribution systems. Appl. Environ. Microbiol. 76 (13) 4169-4178. https://doi.org/10.1128/AEM.03147-09 [ Links ]
LECLERC H, SCHWARTZBROD L and DEI-CAS E (2002) Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28 (4) 371-409. https://doi.org/10.1080/1040-840291046768 [ Links ]
LOKMAN NA, ITHNIN A, YAHYA W and YUZIR M (2020) A brief review on biochemical oxygen demand (BOD) treatment methods for palm oil mill effluents (POME). Environ. Technol. Innov. 21 1-28. https://doi.org/10.1016/Leti.2020.101258 [ Links ]
NAIDOO S and OLANIRAN AO (2014) Treated wastewater effluent as a source of microbial pollution of surface water resources. Int. J. Environ. Res. Public Health 11 (1) 249-270. https://doi.org/10.3390/ijerph110100249 [ Links ]
NGUYEN AQ, VU HP, NGUYEN LN, WANG Q, DJORDJEVIC SP, DONNER E, YIN H and NGHIEM LD (2021) Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 738 146964. https://doi.org/10.1016/j.scitotenv.2021.146964 [ Links ]
MICROCHEM LABORATORY (2021) Log and percentage reductions in microbiology and antimicrobial testing. URL: www.microchemlab.com/information/log-and-percentage-reductions-microbiology-and-antimicrobial-testing (Accessed 20 March 2021). [ Links ]
OMAR KB and BARNARD TG (2010) The occurrence of pathogenic Escherichia coli in South African wastewater treatment plants as detected by multiplex PCR. Water SA. 36 (2) 172-176. https://doi.org/10.4314/wsa.v36i2.183725 [ Links ]
RAMIREZ-CASTILLO FY, LOERA-MURO A, JACQUES M, GARNEAU P, AVELAR-GONZALEZ FJ, HAREL J, and GUERRERO-BARRERA AL (2015) Waterborne pathogens: detection methods and challenges. Pathogens. 4 (2) 307-334. https://doi.org/10.3390/pathogens4020307 [ Links ]
REINTHALER FF, POSCH J, FEIERL G, WÜST G, HAAS D, RUCKENBAUER G, MASCHER F and MARTH E (2003) Antibiotic resistance of E. coli in sewage and sludge. Water Res. 37 (8) 1685-1690. https://doi.org/10.1016/S0043-1354(02)00569-9 [ Links ]
RODRÍGUEZ-MOLINA D, MANG P, SCHMITT H, CHIFIRIUC MC, RADON K and WENGENROTH L (2019) Do wastewater treatment plants increase antibiotic resistant bacteria or genes in the environment? Protocol for a systematic review. Syst. Rev. 8 304. https://doi.org/10.1186/s13643-019-1236-9 [ Links ]
SHEJALE KP, YADAV D, PATIL H, SAXENA S and SHUKLA S (2020) Evaluation of techniques for the remediation of antibiotic-contaminated water using activated carbon. Mol. Syst. Des. Eng. 5 743-756. https://doi.org/10.1039/C9ME00167K [ Links ]
TARR CL, LARGE TM, MOELLER CL, LACHER DW, TARR PI, ACHESON DW and WHITTAM TS (2002) Molecular characterization of a serotype 0121:H19 clone, a distinct shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70 (12) 6853-6859. https://doi.org/10.1128/IAI.70.12.6853-6859.2002 [ Links ]
WHO and UNICEF (World Health Organization and UNICEF) (2017) Progress on Drinking Water, Sanitation and Hygiene: 2017 Update and SDG Baselines. WHO and UNICEF, Geneva. [ Links ]
WHO (World Health Organization) (2018) Drinking water. URL: https://www.who.int/news-room/fact-sheets/detail/drinking-water (Accessed 6 May 2021). [ Links ]
 Correspondence:
Correspondence:
F Tshabuse
Email: freedomvincent@gmail.com
Received: 7 September 2021
Accepted: 7 October 2022
APPENDIX