Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
Water SA
versión On-line ISSN 1816-7950
versión impresa ISSN 0378-4738
Water SA vol.45 no.3 Pretoria jul. 2019
http://dx.doi.org/10.17159/wsa/2019.v45.i3.6730
RESEARCH PAPERS
Physico-chemical properties and bacterial community structure dynamics during the mesophilic anaerobic digestion of pit latrine faecal sludge
MC ChangaraI, *; WT SanyikaII; C BangiraIII; S MisiIV
IDepartment of Environmental Sciences and Technology, Chinhoyi University of Technology, P.O. Box 7724, Chinhoyi, Zimbabwe
IIDepartment of Biotechnology, Chinhoyi University of Technology, P.O. Box 7724, Chinhoyi, Zimbabwe
IIIDepartment of Agricultural Engineering, Chinhoyi University of Technology, P.O. Box 7724, Chinhoyi, Zimbabwe
IVDepartment of Civil Engineering, University of Zimbabwe, P.O. Box MP167, Mount Pleasant, Harare, Zimbabwe
ABSTRACT
The study characterized the changes in physico-chemical properties and bacterial community structure during mesophilic anaerobic digestion (AD) of pit latrine sludge. The sludge was sampled from six different pits six times at an interval of 40 days. Standard techniques were used to assess the changes in pollution indicators including COD and faecal coliforms. Metagenomic DNA from a composite sample from the six pits' sludge was then extracted at Days 0, 14 and 35 and directly sequenced followed by analysis of the microbial structure using the Ribosomal Database Project tools. Multivariate analyses were used to identify the main determinants of microbial community structure during the digestion process. AD significantly reduced the levels of pollution indicators (p < 0.05). Total solids, volatile solids and COD were reduced by 17-27%, 52-79%, and 42-63%, respectively. The indicator pathogenic microorganisms FC and E. coli were reduced by 34-54% and 35-60%, respectively. The reduction in terms of COD and BOD were, however, not sufficient to satisfy the standards for safe disposal into the environment. Proteobacteria were the most dominant bacterial phylum in the undigested sludge (24.1%) and were significantly reduced to 2.5% at the peak of the AD (Day 14) up until Day 35. Firmicutes significantly increased (p < 0.05) from 22.4% to 28.8% at Day 14 before being reduced to 11.6% at Day 35. This study contributes to our understanding of AD of pit latrine faecal sludge through mesophilic AD as a baseline study, and helps to inform future research on mesophilic AD.
Keywords: anaerobic digestion, bacterial community structure, next generation sequencing, pit latrine sludge, pollution indicators
INTRODUCTION
Globally one of the targets of Sustainable Development Goal (SDG) No. 6 is for all nations to provide access to adequate and equitable sanitation and hygiene for all, and to end open defecation by 2030 (WHO/UNICEF, 2017). The drive of improving access to clean drinking water and safe sanitation, according to Montgomery and Elimelech (2007), is one of the least expensive and most effective ways to improve public health and save lives. Hutton et al. (2007) states that improved sanitation has significant economic benefits, for example, the return on 1 USD spent on water and sanitation improvements in low-income countries is 5-46 USD depending on the intervention. The traditional approach when urban and peri-urban houses are being built in developing countries is that every community shall be able to have access to sewer-based systems and the 'flush and forget' stance shall remain forever. Sewer-based systems, however, are usually too costly, too complex and sometimes use too much energy to be implemented in poor and less-developed countries (Lalander et al., 2013; Mara, 2013). Pit latrines have been constructed in peri-urban settlements of developing countries like Zimbabwe as a faecal sludge management technology to substitute sewer-based systems. Pit latrines are among the affordable on-site sanitation facilities (Torendel et al., 2016) in developing countries, but will eventually fill-up at some stage and must be replaced or emptied. Replacement or emptying causes inconvenience is expensive and a health risk (Torendel et al., 2016).
Colon et al. (2015) highlights that one of the greatest challenges with sanitation is developing and managing innovative, user-friendly and easy to adapt low-cost pit faecal sludge disposal systems that are relevant to the local context. In addressing the management of faecal sludge from full latrine pits, there is a growing interest in technologies that not only treat human waste but also enable the recovery of nutrients for agricultural productivity and energy (Gijzen, 2002; Katukiza et al., 2012). However, the pathogenic microorganisms commonly found in human faeces are a cause for concern regarding human health if human excreta are used for crop production. These pathogenic microorganisms include Campylobacter jejuni/coli, E. coli, Salmonella typhi/paratyphi, Salmonella spp., Shigella spp., and Vibrio cholera (Drangert, 1998; Schonning and Stenstrom, 2004). Parasitic microorganisms and worms commonly found in human faecal waste include Cryptosporidium parvum and helminths; Ascaris lumbricoides (roundworm), Taenia solium/saginata (tapeworm), Trichuris trichiura (whipworm), Ancylostoma duodenale/Necator americanus (hookworm) and Schistosoma spp. (blood flukes) (Schonning and Stenstrom, 2004; WHO, 2006).
The process of AD has been found to reduce the levels of pathogens and pollution indicators while retaining plant nutrients (Sahlstrom et al., 2004). AD is a well-established process in which bacteria convert organic wastes to methane and carbon dioxide, a gas mixture (~60% methane and 40% carbon dioxide) called biogas (Lukehurst, 2010). According to Gijzen (2002) and McCarty et al. (2011), the process of AD has a lot of advantages compared to other treatment options, mainly due to low operational costs, production of energy in the form of biogas and potential reuse of the nutrients present in the digested waste. In developing countries such as Ghana the interest in AD is shifting from energy to onsite sanitation systems associated with faecal sludge management, especially in relation to some challenges in emptying, transportation and disposal of pit latrine sludge (Boot and Scott, 2008) and the technology can as well be explored in Zimbabwean context.
Little reliable data is available on the effectiveness of AD of human excreta, both at laboratory and pilot scale (Colon et al., 2015), for agricultural re-use and safe disposal of the digested sludge. Of the little available information on AD of human excreta, much of the emphasis is on energy and biogas production (Park et al., 2001; Colon et al., 2015; Onabanjo et al., 2016). Forbis-Stokes et al. (2016) did a pilot study at two different locations on onsite faecal sludge treatment through AD and recorded 85 and 89% reduction in terms of COD in the effluent in relation to the estimated faecal sludge input. Also log reductions of faecal coliforms recorded in the sterilization tank due to AD were greater than 5. Nwaneri et al. (2008) found that COD values for faecal material present in the first layer of pit latrine sludge were significantly lower than those measured for fresh faeces. The reduction in pollution indicators should, however, meet regulatory standards, for example, the WHO guidelines of 2006 (WHO, 2006), to ensure environmental safety and agricultural utilization of the treated sludge.
In the process of AD, there is also a need to increase the existing knowledge on the dynamics of the complex interacting microbial community that drives the process (Venkiteshwaran et al., 2016). Diverse microbial communities degrade organic matter within pit latrines, and little is known about the specific communities present in pit latrines and their association with faecal decomposition within the pit environment (Torendel et al., 2016). One key area requiring new knowledge involves understanding the effect of AD on the changes in bacterial community structure associated with the pit latrine faecal sludge. The understanding of the microbial community structure and ecology will help in the development of strategies to maintain and improve treatment efficiency (Narihiro et al., 2015).
The aim of this research was to assess the changes in physico-chemical properties and bacterial community structure during the mesophilic AD of pit latrine faecal sludge, and, secondly, to assess the effectiveness of the digestion to treat the faecal sludge to the quality requirements expected for agricultural re-use and safe environmental disposal, for example, the WHO (2006) guidelines for use in crop production and safe disposal. The bacterial community structure and dynamics were investigated through the analysis of the bacterial 16S ribosomal RNA (rRNA) gene diversity generated through Next Generation Sequencing (NGS).
MATERIALS AND METHODS
Sampling of pit latrine sludge
Samples were collected from 6 pit latrines in Shackleton (30.03°E and 17.30°S), located in Chinhoyi, Zimbabwe, during the period February-October 2016, 6 times from each pit at 40-day intervals. Pit latrines were selected purposively, being of approximately 2 years in use, constructed to a depth of approximately 2 m, accessed by more than one family of at least 8 adults (above 16 years) and without connection to water sources. Sampling was conducted using a modified auger which was graduated to show sampling depth. After determining the depth of each pit latrine, triplicate faecal samples were taken from each pit (about 50-60 cm pit slab to contents depth). The sludge was sampled from the top surface about (10 cm deep) of the pit latrine sludge and the modified auger that was used for sampling collected about 135 g of sludge at each time. Composite samples of approximately 275 g were placed in 300 mL clear polythene bottles for AD, physico-chemical characterisation and microbial composition analysis. A total of 2 kg moist sludge was collected per each pit at each sampling time. The samples were immediately placed on ice and later transported to the laboratory for physico-chemical, microbial analysis and AD. Physico-chemical and microbial analysis excluding metagenomic DNA analyses were conducted after addition of 10% (w/v) rumen fluid inoculums before and after the AD for the 6 sampling times per pit. A composite sample was then collected from all 6 pits for which metagenomic DNA and physico-chemical properties were determined at Day 0, both without addition of rumen and after addition of rumen, at the climax of biogas production (Day 14) and at the end of the AD (Day 35). The sampling points for the metagenomic analysis were determined from preliminary experiments.
Inoculum preparation
A 150 mL volume of rumen fluid with a total solid content of 7% was obtained from a nearby abattoir (The Cold Storage Commission, Chinhoyi) and used as inoculum as prescribed by Ojolo et al. (2007). The rumen fluid was first mixed with distilled water in a ratio 1:1 (v/v). A 5-mm sieve was used to separate solid content from liquid in the preparation of the inoculum.
Sample preparation and anaerobic digestion
A 1.5 kg moist faecal pit latrine sludge sample from each of the 6 pits (total solids content ranging from 3 to 7%, depending on different pits and different sampling times) was fed into the 2-L high-density polyethylene (HDPE) plastic container reactors (Fig. 1). Each reactor was fitted with a flexible 12 mm gas delivery pipe (Fig. 1) and jacketed with cotton wool to insulate the internal working temperature from ambient fluctuations. An airtight seal was then used for anaerobic conditions to prevail. The reactors were shaken by hand at least 2 times a day for 30 s. The biogas produced in the reactors was collected in an inverted measuring cylinder in a plastic beaker (Fig. 1) containing 5% sodium hydroxide solution. The amount of biogas produced was measured through the displacement of the sodium hydroxide in the inverted measuring cylinder. At the end of the measuring cylinder there was a 12 mm gas delivery pipe linked to a gas valve then a Bunsen burner (Fig. 1) at the end of the anaerobic digester set up. At the end of each day the cumulative biogas in the inverted measuring cylinder in each reactor was released via the gas valve and a burning test was conducted, checking if the gas produced was burning and producing a blue flame on the Bunsen burner. The cumulative production of burnable biogas was used as a measure of better performance of the reactors in addition to the measurement of changes in physico-chemical properties during the digestion. The experiment was carried out in a batch reaction mode for 35 days at mesophilic temperature (18-35°C).
Analytical methods
Laboratory analyses of total solids (TS), volatile solids (VS), chemical oxygen demand (COD), biological oxygen demand (BOD), total coliforms (TC), faecal coliforms (FC), Escherichia coli (E. coli), helminth eggs, total nitrogen (N), phosphorus (P), potassium (K), calcium (Ca), magnesium (Mg) and sodium (Na) were measured in triplicate before and after a 35-day mesophilic AD in all the digesters for the 6 sampling times. The total chemical nutrients (N, P, K, Ca, Mg and Na) and the physico-chemical parameters (TS, VS, COD and BOD) were determined according to APHA/AWWA/WEF (2005) and Reddy (2013). Total coliforms, faecal coliforms and E. coli were determined using a Most Probable Number assay (MPN) method 8001A (USEPA, 2012). Helminth egg determination was done through the modified Bailenger method (Ayres et al., 1996).
DNA extraction
The DNA was extracted from 150 mg of each sample using the ZR Fecal DNA MiniPrep kit (Zymo research) following the manufacturer's instructions. The DNA was extracted from Day 0, Day 14 and Day 35 sludge samples during AD.
Polymerase chain reaction (PCR) and next generation sequencing (NGS)
The V1 and V3 variable regions were amplified in a 25 µL reaction using Q5 Hot start High-Fidelity 2x Master Mix (New England Biolabs, USA). Amplicon library PCR was performed on all replicate extractions separately. The DNA primers used were 27F-16S AGAGTTTGATCMTGGCTCAG, 518R-16S ATTACCGCGGCTGCTGG. Thermal cycler setting for PCR amplification were as follows: (i) initial denaturation at 95°C for 2 min, (ii) 30 cycles of 95°C for 20 s, (iii) 55°C for 30 s, (iv) 72°C for 30 s and final elongation at 72°C for 5 min. The amplicon libraries were purified using the Agencourt AMPure XP bead protocol (Beckman Coulter, USA). Library concentration was measured using Nebnext Library quant kit (New England Biolabs, USA) and quality validated using Agilent 2100 Bio analyser (Agilent Technologies, USA). The samples were pooled in equimolar concentrations and diluted to 4nM based on library concentrations and calculated amplicon sizes. The library pool was sequenced on a MiSeq (illumina, USA) using a MiSeq Reagent kit V3 600 cycle PE (illumina, USA). The final pooled library was at 12 pM with ١٥٪ PhiX as control. The illumina MiSeq automatically detected the barcodes and subsequently demultiplexed the clusters that passed on-board quality filter checks and the output was formatted as fastq files.
Analysis of pyrosequencing-derived data
Sequence quality checks were analysed using the Ribosomal Database Project (RDP) pipeline. The sequences were cleaned using the RDP GL FLX software Release 11 (http://rdp.cme.msu.edu/) (Cole et al., 2014), with the following filters minimum read Q score of 30, minimum sequence length of 150 bp and maximum sequence length of 500 bp. Raw sequence reads were filtered before subsequence analyses to minimize the effect of random sequencing errors. The pipeline initial process was used to remove sequences of low quality.
After the initial processing, the high-quality sequences were then assigned to the bacterial taxonomy using the RDP Classifier (Wang et al., 2007) at 80% confidence interval. Classification of the newly generated sequences was based on the trained RDP Naive Bayesian rRNA Classifier (Version 2.2, March 2010) of the RDP (Wang et al., 2007). Sequence alignment of the effective bacterial sequences of each sample was done using fast, secondary-structure aware Infernal aligner version 1.1rc4 (Nawrocki et al., 2009). After sequence alignment an aligned file was obtained from each sample. The aligned files were used to cluster the sequences of each sample using the RDP Complete Linkage Clustering tool and a cluster file obtained. The comparison of the microbial communities between Day 0, Day 14 and Day 35 was conducted using the RDP library compare tool at 80% confidence interval.
Statistical analysis
The software, Primer 7 (Primer-E) version 7.0.13 (Quest Research Limited, Auckland, New Zealand) and Multivariate Statistical Package (MVSP) version 3.1 (KOVACH Computing Services, United Kingdom) were used for statistical analyses. Hierarchical clustering and multidimensional scaling (MDS) were based on a similarity matrix generated from standardized and normalized data. The Euclidean distance was used for environmental data. The hierarchical clustering was conducted using the group average method. Multidimensional scaling ordinations were based on 10 iterations and cluster overlays were based on cluster analysis. ANOSIM (analysis of similarity) was used to test the strength of separation and for significant differences between the different clusters and pit latrine sludge samples. Principle component analysis (PCA) was preceded by a distance matrix of the dataset. MVSP was used for canonical correspondence analysis (CCA). A paired t-test was performed to test for significant differences between undigested and anaerobically digested pit latrine sludge physico-chemical and microbial parameters per pit per cycle of digestion.
RESULTS AND DISCUSSION
Bioreactor performance
The temperature in all the digesters ranged between 23°C and 34°C (Fig. 2a). The highest temperature was recorded at Days 14 and 15 and the lowest temperature in all digesters was recorded at the first and the last days of the digestion process in all the digesters. The temperature remained at mesophilic range throughout the study while pH fluctuated between 8.1 and 5.1 in all the digesters (Fig. 2c). Generally, the pH decreased from the beginning of the digestion and the lowest pH values were recorded at Days 13 to 16 before increasing again until Day 35 (Fig. 2c). The COD values decreased at a faster rate, mainly between Day 0 and Day 14 and thereafter continued to decrease up to Day 35 but at a much lower rate (Fig. 2b). The general trend of COD was similar to that observed for BOD, TS, and VS.
The cumulative volume of biogas produced from the different pit latrine sludge digesters ranged from 0.001 to 0.003 m3/KgVSa with maximum daily production observed at Day 14 (Fig. 2d). The biogas ignited in all reactors with the earliest combustion recorded on Day 6.
General trends of physico-chemical properties on net effect of AD on pit latrine sludge quality
Based on nMDS analysis (data not shown) the undigested sludge samples were classified separately from treated samples, irrespective of the space (pit) and time (sampling period) in which the samples were collected. The physico-chemical properties of treated and untreated sludge were distinct enough to be classified into these separate categories irrespective of pit or sampling period. The PCA confirmed the nMDS result that undigested sludge samples classified separately from treated samples and showed that TS, VS, COD and BOD mostly accounted for the variability between the digested and undigested sludge treatments (Fig. 3).

The process of AD resulted in changes in physico-chemical properties as highlighted by the changes in pH and COD (Fig. 2). Cluster analysis (data not shown), nMDS and ANOSIM (data not shown) results also indicated that the mesophilic AD of pit latrine sludge resulted in significant changes in physico-chemical characteristics of the sludge. Biogas production (Fig. 2d) confirmed that the process of AD was complete in each digester.
Effect of anaerobic digestion on selected individual pollution indicators
At the level of individual pollution indicators, AD significantly reduced COD and BOD (p < 0.05) in all the pit latrine sludge samples (Table A1, Appendix). The percentage reduction for the COD and BOD ranged from 42-63% and 34-52%, respectively (Table A1, Appendix). These are comparable to the values of 58-61% reported by Issah et al. (2012) and Marti et al. (2008). The COD concentrations in the digested sludge from all the digesters, which ranged from 38 733 ± ١٠ 452 to 53 563 ± 6 875 mg/L (Table A1, Appendix) were, however, still above the environmental safety disposal requirements of the Zimbabwe Regulations (Government of Zimbabwe, 2007) of < 60 mg/L of COD by approx. 3 orders of magnitude. Thus the digested pit latrine sludge cannot be disposed into the environment without further treatment. The BOD strength of the anaerobically digested pit latrine sludge, which ranged from 8 048 ± ١ 179 to 10 916 ± 743.9 mg/L, also remained higher than the WHO BOD5 (1989) standard of 20-100 mg/L for agricultural use and disposal to the environment.
Feacal coliforms (FC) and E. coli were also significantly reduced (p < 0.05) by the process of AD (Fig. A1). The percentage reductions of FC and E. coli ranged from 33-61% in all the samples (Table B.1). Helminth eggs were not detected in any of the pit latrine sludge samples, both before and after AD. The changes in ecological conditions during the AD process, including temperature and pH, eliminate the sensitive bacteria, including coliforms (Pandey and Soupir, 2011). In comparison, other studies have reported 50-70% reduction in pathogens using AD under mesophilic conditions (Sunders and Harrison, 2011; Issah et al., 2012), which was slightly higher than the percentage reduction in this study.
Although significant reduction of FC occurred in each digester during AD, the number of FCs in the digested sludge, which ranged from 1.0 x 106 ± ٢.٧ x ١٠4 to 2 x 106 ± 1.8 x 105 MPN/100 mL, still exceeded the environmental safety disposal limit of <1 000/100 mL imposed by the Zimbabwe Regulations (Government of Zimbabwe, 2007).
Overall, the process of AD, as expected, significantly reduced the concentration and the counts of pollution indicators (COD, BOD, FC and E. coli) in the undigested sludge in comparison to the digestate. The concentration and the counts of the pollution indicators (COD and BOD) in the digestate were, however, still above the WHO (2006) guidelines for re-use of the waste in agriculture. It has to be acknowledged that the process of mesophilic AD, in as much as it significantly reduces pollution indicators in the pit latrine sludge, did not reduce these to levels acceptable for re-use or safe disposal. The process of mesophilic AD of pit latrine sludge on its own cannot thus be recommended for treatment of pit latrine sludge for safe disposal. The main thrust of this research was, however, to determine the extent to which (in terms of percentage reduction) the process of mesophilic AD reduces pollution indicators in pit latrine sludge, as a first step (baseline research) towards treating pit latrine sludge and to inform further studies.
Bacterial community structure changes during AD
Estimates of diversity
A total of 15 087, 7 683 and 9 715 pyrosequences were generated at Day 0, Day 14 and Day 35, respectively (Table 1). After sorting and trimming processes, a total of 11 478, 5 880 and 7 690 sequences of acceptable quality were obtained for Day 0, Day 14 and Day 35, respectively. These sequences, with an average length of 293-295 bp, were used to construct 16S rRNA gene libraries for subsequent analysis.
Diversity indices provide more information about species representation in community composition than just species richness. In this study the Shannon-Weaver Index (H) and the Chao 1 estimator were used to estimate the statistical parameters that described each community structure. The analysis of evenness (E) was based on the OTU data set. In general, all the samples had balanced populations that had high diversity indices, evenness and species richness. It was difficult to compare the diversity parameters in this study with other studies because of the differences in the reads generated from the pyrosequencing.
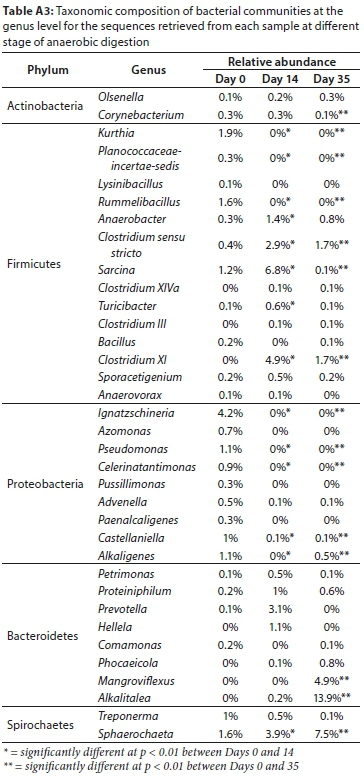
A total of 9 bacterial phyla (Fig. 4) were identified in the undigested pit latrine sludge that was inoculated with 10% (v/v) cow rumen fluid. The dominant bacteria in the undigested pit latrine sludge inoculated with rumen were of the phylum Proteobacteria 26.5%, then Firmicutes 25.1% followed by Spirochaetes (4.7%), Bacteroidetes 2.2% and Actinobacteria 0.9%. The rest of the bacterial phyla included the Planctomycetes, Synergistestes, Verrucomicrobia and Lentisphaearae, which were underrepresented (Fig. 4).

Comparison of microbial community composition clearly showed how this varied between the anaerobically digested sludge and the undigested samples. The Bacteroidetes increased from 2.2% at Day 0 to 16.3% at Day 14 and 32.1% at Day 35. Firmicutes increased from 22.4% at Day 0 to 28.8% at Day 14 before decreasing to 11.6% at Day 35. The Spirochaetes increased from 4% at Day 0 to 5.5% at Day 14 and further increased to 9.2% at Day 35. Proteobacteria decreased significantly to 2.8% at Day 35 from 24.1% at Day 0 to 2.5% at Day 14. All the changes were significant at p < 0.05.
It must be acknowledged that the bacterial composition reported here was after the addition of 10% cow rumen which could affect sludge bacterial composition. However, a comparison of the pit latrine sludge alone and that mixed with 10% cow rumen (Table 2) shows that the percentage distribution of the dominant phyla was the same.

A comparison of the bacterial phyla in this study after the addition of rumen with other pit latrine sludge samples and with rumen bacterial composition from literature suggest that the trends of the bacterial composition in this study follow that of the pit latrine sludge rather than cow rumen, though the contribution of the rumen fluid cannot be overruled. For example, Proteobacteria represented 26.5% of the bacterial community in our study, and, in another study, 25% in pit latrine sludge samples from Vietnam (Torendel et al., 2016), whereas a study by Ozbayram et al. (2018) found no Proteobacteria in in cattle rumen. Ozbayram et al. (2018) highlighted that the dominant phyla in cattle rumen were Bacteroidetes (54%); in this study Bacteroidetes contribution was 2.2%. In the same study of cattle rumen by Ozbayram et al. (2018), Fibrobacteres and Lentisphaerae contributed 12% and 8%, respectively, but these phyla were not present or underrepresented in our study as well as in other pit latrine sludge samples from Tanzania and Vietnam (Torendel et al., 2016).
The process of AD was characterised by changes in community structure that were associated with biochemical and physico-chemical changes. These changes are usually attributed to the processes of hydrolysis, acidogenesis, acetogenesis and methanogenesis. The microbial diversity observed in the undigested sludge (Day 0) are associated with the early stages of AD and minimal activity in terms of biogas production. The activity (biogas production) increased reaching a peak at Day 14 which also corresponded with the highest temperature reached on this day (Fig. 2). Day 14 was therefore associated with maximum activity or maximum cumulative activity and the turning point towards the decline of activity (biogas production and temperature) during the anaerobic digestion (Fig. 2). Day 35 presented the latter stages of the process, which were characterised by declining and almost stationary biogas production (Fig. 2).
Our findings indicated that Proteobacteria, Firmicutes, Spirochaetes, Bacteroidetes and Actinobacteria were the major bacterial phyla commonly found in fresh faecal sludge in the early stages of digestion that occurs in the uppermost layer of pit latrines. The bacterial composition can be altered by a number of environmental factors, including anal cleansing and bathing material, and the design of the latrine, amongst other factors (Torendel et al., 2016). The Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria have been reported to be the most abundant phyla in pit latrine sludge bacterial communities (McLellan et al., 2010; Torendel et al. 2016). Sewage sludge profiles were also found to be dominated by the same phyla (Shanks et al., 2013). Firmicutes are common inhabitants of the human gut (Gill et al., 2006) and environments where AD occurs like activated sludge systems and anaerobic digesters (Garcia-Pena et al., 2011). One of the dominant genera, Treponerma (Table A3, Appendix), is from the phylum Spirochaetes, which have previously been shown to be important components of the gut microbiota in individuals from rural, less developed regions (Schnorr et al., 2014), which is relevant to this study as the sludge was collected from a peri-urban area which is considered to be a poor area, for example, in terms of diet. Proteobacteria are important microbes in the AD process because most of the Alpha-, Beta-, Gamma-, and Deltaproteobacteria are well known glucose, propionate, butyrate, and acetate-utilizing microbial communities (Ariesyady et al., 2007).
As the AD process occurred, there were notable changes in numbers of the inherently emerging bacterial populations and disappearance of those involved in the early stages of AD, showing that the process involves microbial succession and specific groups of microorganisms drive the specific biotransformations of sludge material towards completion. A shift in this succession could therefore be detrimental to the efficient production of biogas or effective treatment of the faecal sludge. Understanding these changes in microbial community structure is therefore very important for monitoring and improving the efficiency of AD of faecal sludge and derivation of maximum value. The turning point of activity, Day 14, was characterised by drastic shifts of bacteria at phylum level, with an increase in Firmicutes of the class Clostridia, Spirochaetes of the class Spirochaetea and Bacteroidetes of the classes Bacteroidia and Flavobacteria. These results suggest that an increase in numbers occurred in those bacterial groups actively involved in the turning point of the process or simply those adaptable to the emerging conditions. Most members of Firmicutes are syntrophic bacteria that can degrade various volatile fatty acids (Garcia-Pena et al., 2011). The Bacteroidetes, which increased up to Day 14, are known to produce various lytic enzymes and acetic acid during the degradation of organic materials (Chen and Dong, 2005; Robert et al., 2007; Riviere et al., 2009).
At Day 35, which was considered the end of the digestion process, there was a reduction in Firmicutes of the class Bacilli and further reduction in Proteobacteria of classes Alphaproteobacteria, Gammaproteobacteria and Betaproteobacteria. This reduction in numbers found in the undigested sludge was an indication of those bacteria that were reduced or eliminated by the process of AD or that did not favourably respond to the changes in conditions during the process.
The results obtained in this study in terms of bacterial shifts during AD was comparable to those of Alcantra-Hernandez et al. (2017) and Doloman et al. (2017), who reported that Proteobacteria diminished from 33% to 10-16% whilst Spirochaetes and Bacteroidetes increased as a result of AD. Their results were, however, different in relation to Firmicutes; we observed a significant decrease in Firmicutes as a result of AD, whilst Alcantra-Hernandez et al. (2017) and Doloman et al., (2017) reported an increase of Firmicutes as a result of AD. The waste material for the study by Alcantra-Hernandez et al. (2017) was organic urban residues and for that of Doloman et al. (2017) was microbial biomass, both being operated at mesophilic conditions.
Association between bacterial community structure and environmental properties at Days 0, 14 and 35 of AD
Using canonical correspondence analysis (CCA) at genus level. Treponerma, Alkaligenes and Celerinatantimonas were associated with high COD, BOD and VS at Day 0. Prevotella, Hellela, and Sarcina were associated with high production of biogas at Day 14, which was the turning point for all the measured physico-chemical properties. Mangroviflexus, Alkalitalea and Oligosphaera were associated with high K and low COD, BOD and VS at Day 35 (Fig. 4).

It should be appreciated that no two pits are alike; even the same pit sampled at different depths is not alike (Bakare et al., 2012). What we have assessed here is a general trend from specific pit latrine sludge in Shackleton, but specific details could differ from pit to pit and area to area. Factors such as differences in gut microbiota, diet, anal cleansing material and pit latrine maintenance approaches, like different emptying patterns, can result in differing microbial compositions of raw pit latrine sludge (Torendel et al., 2016). The results presented here were obtained from small-scale reactors and thus may not exactly replicate the much bigger digesters and as such the hydraulic retention times, reactor temperatures, and mixing methods will likely require modifications during scale up. However, the results obtained using this anaerobic digester model provides insights into how full-scale reactors may perform.
To the best of our knowledge this is the first study describing the changes in microbial community structure during AD of pit latrine sludge. Most studies have traditionally been based on culture-dependent techniques targeting specific bacterial groups such as E. coli and faecal coliforms, which are indicators of faecal origin. The use of next generation sequencing based on the 16S rRNA identification reveals diversity that would not have been shown by culture dependant techniques. The findings from this study indicate the key bacterial groups involved in the AD process, which could be useful for improving the efficiency of the AD process and developing bacteria-based technological solutions for the treatment of pit latrine sludge and energy generation from the sludge.
CONCLUSIONS
The study revealed that the process of mesophilic AD of pit latrine sludge reduces pollution indicators in the sludge. The reduction was, however, insufficient to meet the threshold for safe disposal. The process of mesophilic AD of pit latrine sludge on its own cannot therefore be recommended for treatment of pit latrine sludge for safe disposal. The study, however, has provided information on qualitative and quantitative changes in the levels of pollution indicators during the AD process. The study also showed that there is wide diversity of bacteria in pit latrine sludge, which changes in community structure as the AD process progresses. The changes in community structure described by this study are considered to be the changes that occur during healthy mesophilic AD of faecal pit latrine sludge. The process may, however, be affected by a number of factors that may alter the succession reported here and subsequently affect the efficiency of the process. The process of mesophilic AD promotes proliferation of Bacteroidetes and Spirochaetes and significantly reduces Proteobacteria and Firmicutes. The information generated will form a basis for future research in understanding the process of mesophilic AD of pit latrine sludge. The information generated can also be used to compare mesophilic AD with other proposed treatment options for pit latrine sludge and could be used to develop microbial-based treatment technologies and management solutions.
ACKNOWLEDGEMENTS
The authors would like to thank the Sanitation Research Fund for Africa (SRFA) for funding this research project through the Water Research Commission (SA), and Chinhoyi University of Technology for providing laboratories to carry out the experiments and allowing for the research to be carried out on campus.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
ALCANTARA-HERNANDEZ RJ, TAS N, CARLOS-PINEDO S, DURAN-MORENO A and FALCON LI (2017) Microbial dynamics in anaerobic digestion reactors for treating organic urban residues during the start-up process. Lett. Appl. Microbiol. 64 438-445. https://doi.org/10.1111/lam.12734 [ Links ]
APHA/AWWA/WEF (2005) Standard Methods for the Examination of Water and Wastewater, 20th edition. American Public Health Association/American Water Works Association/Water Environment Federation, Washington DC, USA. https://doi.org/10.12999/awwa.g510.13 [ Links ]
AYRES RM, MARA DD and RACHEL M (1996) Analysis of Wastewater for Use in Agriculture - A Laboratory Manual of Parasitological and Bacteriological Techniques. World Health Organisation, Geneva, Switzerland. 1-35. URL: http://www.who.int/iris/handle/10665/41832 (Accessed 3 February 2015). [ Links ]
ARIESYADY HD, ITO T and OKABE S (2007) Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 41 1554-1568. https://doi.org/10.1016/j.watres.2006.12.036 [ Links ]
BAKARE BF, FOXON KM, BROUCKAERT CJ and BUCKLEY CA (2012) Variation in VIP latrine sludge contents. Water SA 38 479-486. https://doi.org/10.4314/wsa.v38i4.2 [ Links ]
BOOT NLD and SCOTT RE (2008) Faecal sludge management in Accra Ghana: strengthening links in the chain. In: Proceedings of the 33rd WEDC International Conference - Access to sanitation and safe water global partnerships and local actions, 7-11 April 2018, Accra, Ghana. https://doi.org/10.3362/1756-3488.2008.007 [ Links ]
CHEN S and DONG X (2005) Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 55 (6) 2257-2261. https://doi.org/10.1099/ijs.0.63807-0 [ Links ]
COLE JR, WANG JA, FISH B, CHAI DM, MCGARRELL Y, SUN CT, BROWN A, PORRAS-ALFARO C, KUSKE R and TIEDJE JM (2014) Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucl. Acids Res. 42 (Database issue) D633-D642. https://doi.org/10.1093/nar/gkt1244 [ Links ]
COLON J, FORBIS-STOKES AA and DESHUSSES MA (2015) Anaerobic digestion of undiluted simulant human excreta for sanitation and energy recovery in less-developed countries. Energ. Sustain. Dev. 29 57-64. https://doi.org/10.1016/j.esd.2015.09.005 [ Links ]
DRANGERT JO (1998) Fighting the urine blindness to provide more sanitation options. Water SA 24 (2) 157-164. [ Links ]
DOLOMAN A, SOBOH Y, WALTERS AJ, SIMS RC and MILLER CD (2017) Qualitative analysis of microbial dynamics during anaerobic digestion of microalgal biomass in a UASB reactor. Int. J. Microbiol. (2017) 1-12. https://doi.org/10.1155/2017/5291283 [ Links ]
FORBIS-STOKES AA, O'MEARA PF, MUGO W, SIMIYU GM and DESHUSSES MA (2016) On-site fecal sludge treatment with the anaerobic digestion pasteurization latrine. Environ. Eng. Sci. 33 (11) 898-906. https://doi.org/10.1089/ees.2016.0148 [ Links ]
GARCIA-PENA EL, PARAMESWARAN P, KANG DW, CANUL-CHAN M and KRAJMALNIK-BROWN R (2011) Anaerobic digestion and co-digestion processes of vegetative and fruit residues: Process and microbial ecology. Bioresour. Technol. 102 9447-9455. https://doi.org/10.1016/j.biortech.2011.07.068 [ Links ]
GIJZEN HJ (2002) Anaerobic digestion for sustainable development: A natural approach. Water Sci. Technol. 45 (10) 321-328. https://doi.org/10.2166/wst.2002.0364 [ Links ]
GILL SR, POP M, DEBOY RT, ECKBURG PB, TURNBAUGH PJ, SAMUEL BS, GORDON JI, RELMAN DA, FRASER-LIGGETT CM and NELSON KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312 1355-1359. https://doi.org/10.1126/science.1124234 [ Links ]
GOVERNMENT OF ZIMBABWE (2007) Environmental Management (Effluent and Solid Waste Disposal) Regulations (2007) Supplement to the Zimbabwean Government Gazette dated 5th of January, 2007. Government Printer, Harare. [ Links ]
HUTTON G, HALLER L and BARTRAM J (2007) Global cost-benefit analysis of water supply and sanitation interventions. J. Water Health 5 (4) 481-502. https://doi.org/10.2166/wh.2007.009 [ Links ]
ISSAH A, AKLAKU ED and SALIFU T (2012) Comparative study of effluent for pollution indicators and indicator pathogenic organisms from anaerobic digesters from human and fruit wastes. ARPN J. Agric. Biol. Sci. 7 416-419. [ Links ]
KATUKIZA AY, RONTELTAP M, NIWAGABA CB, FOPPEN JW, KANSIIME F and LENS PN (2012) Sustainable sanitation technology options for urban slums. Biotechnol. Adv. 30 (5) 964-978. https://doi.org/10.1016/j.biotechadv.2012.02.007 [ Links ]
LALANDER CH, HILL GB and VINNERAS B (2013) Hygienic quality of faeces treated in urine diverting vermicomposting toilets. Waste Manage. 33 (11) 2204-2210. https://doi.org/10.1016/j.wasman.2013.07.007 [ Links ]
LUKEHURST CT, FROST P and AL SEADI T (2010) Utilization of digestate from biogas plants as bio-fertilizer. IEA Bio-Energy 5-16. [ Links ]
MARA D (2013) Pits, pipes, ponds-And me. Water Res. 47 (7) 2105-2117. https://doi.org/10.1016/j.watres.2013.01.051 [ Links ]
MARTI N, BOUZAS A, SECO A and FERRER J (2008) Struvite precipitation assessment in anaerobic digestion processes. Chem. Eng. J. 141 67-74. https://doi.org/10.1016/j.cej.2007.10.023 [ Links ]
MCCARTY PL, BAE J and KIM J (2011) Domestic wastewater treatment as a net energy producer-can this be achieved? Environ. Sci. Technol. 45 (17) 7100-6. https://doi.org/10.1021/es2014264 [ Links ]
MCLELLAN SL, HUSE S M, MUELLER-SPITZ SR, ANDREISHCHEVA EN and SOGIN ML (2010) Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol 12 (2) 378-392. https://doi.org/10.1111/j.1462-2920.2009.02075.x [ Links ]
MONTGOMERY MA and ELIMELECH M (2007) Water and sanitation in developing countries: including health in the equation. Environ. Sci. Technol. 41 (1) 17-24. https://doi.org/10.1021/es072435t [ Links ]
NAWROCKI EP, KOLBE DL and EDDY SR (2009) Infernal 1.0: inference of RNA alignments. Bioinformatics 25 (10) 1335-1337. https://doi.org/10.1093/bioinformatics/btp157 [ Links ]
NARIHIRO T, KIM N. K, MEI R, NOBU MK and LIU WT (2015) Microbial community analysis of anaerobic reactors treating soft drink wastewater. PLoS ONE 10 (3) e0119131. https://doi.org/10.1371/journal.pone.0119131 [ Links ]
NWANERI CF, FOXON K, BAKARE BF and BUCKLEY C (2008) Biological degradation processes within a pit latrine. In: Proceedings of WISA 2008 Conference, Sun City, South Africa, 19-22 May 2008. [ Links ]
ONABANJO T, KOLIOS AJ, PATCHIGOLLA K, WAGLAND ST, FIDALGO B, JURADO N, HANAK DP, MANOVIC V, PARKER A, MCADAM E, WILLIAMS L, TYRREL S and CARTMELL E (2016) An experimental investigation of the combustion performance of human faeces. Fuel 184 780-791. https://doi.org/10.1371/journal.pone.0119131 [ Links ]
OJOLO SJ, OKE SA, ANIMASAHUM K and ADESUYI BK (2007) Utilization of poultry, cow and kitchen wastes for biogas production: a comparative analysis. Iran J. Environ. Health Sci. Eng. 4 (4) 223-228. [ Links ]
OZBAYRAM EG, INCE O, INCE B, HARMS H and KLEINSTEUBER S (2018) Comparison of rumen and manure microbiomes and implications for the inoculation of anaerobic digesters. Microorganisms 2018 (6) 15. https://doi.org/10.3390/microorganisms6010015 [ Links ]
PARK JA, HUR JM, SON BS and LEE JH (2001) Effective treatment of night soil using anaerobic sequencing batch reactor (ASBR). Korean J. Chem. Eng. 18 (4) 486-492. https://doi.org/10.1007/bf02698295 [ Links ]
PANDEY PK and SOUPIR ML (2011) Escherichia coli inactivation kinetics in anaerobic digestion of dairy manure under moderate mesophilic and thermophilic temperature. AMB Express 1 18. https://doi.org/10.1186/2191-0855-1-18 [ Links ]
REDDY M (2013) Standard Operating Procedures. Howard College, School of Chemical Engineering; Population Research Fund. University of KwaZulu-Natal, Durban. [ Links ]
RIVIERE D, DESVIGNES V, PELLETIER E, CHAUSSONNERIE S, GUERMAZI S, WEISSENBACH J, LI T, CAMACHO P and SGHIR A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 3 (6) 700-714. https://doi.org/10.1038/ismej.2009.2 [ Links ]
ROBERT C, CHASSARD C, LAWSON PA and BERNALIER-DONADILLE A (2007) Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int. J. Syst. Evol. Microbiol. 57 (7) 1516-1520. https://doi.org/10.1099/ijs.0.64998-0 [ Links ]
SAUNDERS O and HARRISON J (2011) Pathogen Reduction in Anaerobic Digestion of Manure. URL: http://www.extension.org/pages/30309/pathogen-reduction-in-anaerobic-digestion-of-manure (Accessed 5 February 2016). [ Links ]
SAHLSTROM L, ASPAN A, BAGGE E, DANIELSSON-THAM ML and ALBIHN A (2004) Bacterial pathogen incidences in sludge from Swedish sewage treatment plants. Water Res. 38 (8) 1989-1994. https://doi.org/10.1016/j.watres.2004.01.031 [ Links ]
SCHNORR SL, CANDELA M, RAMPELLI S, CENTANNI M, BASAGLIA G, TURRONI S, BIAGI E, PEANO C, SEVERGNINI M, FIORI J and co-authors (2014) Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5 (3654) 1-12. https://doi.org/10.1038/ncomms4654 [ Links ]
SCHONNING C and STENSTROM TA (2004) Guidelines for the safe use of urine and faeces in ecological sanitation systems. EcoSanRes Publications Series Report 2004-1, Stockholm Environment Institute, Stockholm. [ Links ]
SHANKS OC, NEWTON RJ, KELTY CA, HUSE SM, SOGIN ML and MCLELLAN SL (2013) Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl. Environ. Microbiol. 79 2906-2913. https://doi.org/10.1128/aem.03448-12 [ Links ]
TORONDEL B, ENSINK JH, GUNDOGDU O, IJAZ UZ, PARKHILL J, ABDELAHI F, NGUYEN VA, SUDGEN S, GIBSON W, WALKER AW and co-authors (2016) Assessment of the influence of intrinsic environmental and geographical factors on the bacterial ecology of pit latrines. Microb. Biotechnol. 9 (2) 209-223. https://doi.org/10.1111/1751-7915.12334 [ Links ]
USEPA (United States Environmental Protection Agency) (2012) Method 8001A: Total Coliforms, Faecal Coliforms and Escherichia coli in wastewater by Most Probable Number Method. URL: http://www.epa.gov/microbes/1604sp02.pdf (Accessed 23 January 2017). [ Links ]
VENKITESHWARAN K, BOCHER B, MAKI J and ZITOMER D (2016) Relating anaerobic digestion microbial community and process function. Water Microbiol. 8 (S2) 37-44. https://doi.org/10.4137/mbi.s33593 [ Links ]
WANG QG, GARRITY M, TIEDJE JM and COLE JR (2007) Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73 (16) 5261-5267. https://doi.org/10.1128/aem.00062-07 [ Links ]
WHO (2006) Guidelines for the Safe Use of Wastewater, Excreta and Grey Water. World Health Organisation, Geneva. [ Links ]
WHO (1989) Health guidelines for the use of wastewater in agriculture and aquaculture. Report of a World Health Organization Scientific Group, WHO Technical Report Series 778. World Health Organization, Geneva. https://doi.org/10.1159/isbn.978-3-318-05724-9 [ Links ]
WHO/UNICEF (2017) Progress on Drinking Water, Sanitation and Hygiene-2017 update and SDG Basilines. WHO/UNICEF, Geneva. [ Links ]
Received 6 June 2018
Accepted in revised form 25 June 2019
* Corresponding author, email: chrischangara@gmail.com
APPENDIX