Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Water SA
On-line version ISSN 1816-7950
Print version ISSN 0378-4738
Water SA vol.38 n.5 Pretoria Jan. 2012
SHORT COMMUNICATION
Isolation and identification of bacterial pollutants from the Berg and Plankenburg Rivers in the Western Cape, South Africa
AN PaulseI; VA JacksonI; S KhanI W KhanII, III, *
IDepartment of Biomedical Sciences, Faculty of Health and Wellness Sciences, Cape Peninsula University of Technology, Bellville 7535, South Africa
IIDepartment of Agricultural and Food Sciences, Faculty of Applied Science, Cape Peninsula University of Technology, Cape Town 8000, South Africa
IIIDepartment of Microbiology, Faculty of Science, Stellenbosch University, Stellenbosch 7600, South Africa
ABSTRACT
Bacterial species present in the Berg and Plankenburg Rivers (Western Cape, South Africa) were isolated from water and biofilm samples and population shifts between sampling sites were phylogenetically identified. Deoxyribonucleic acid (DNA) extraction of representative isolates was performed and amplified using 2 different primer sets. Various Enterobacteriaceae species were present at all of the sites, confirming faecal contamination. Phylogenetic analyses also showed that, in general, Gram-negative micro-organisms dominated at all of the sites sampled in both the Berg and Plankenburg river systems. Pathogens and opportunistic pathogens, such as Pseudomonas aeruginosa, Staphylococcus sp., and Bacillus cereus, were isolated from the Berg River. Similarly, in the Plankenburg River system, Aeromonas sp., Acinetobacter sp., Stenotrophomonas sp. and Yersinia enterocolitica were also isolated. This raises major health concerns as human population densities along both rivers are high, thus resulting in increased human exposure to these organisms.
Keywords: Enterobacteriaceae, faecal contamination, informal settlement, river water, waterborne illnesses
Introduction
South Africa's water resources service various industrial, agricultural and domestic sectors, which in turn may contribute to the point or non-point source contamination of these valuable water sources. In many areas of South Africa, and worldwide, informal housing schemes are often established along the banks of river systems. Due to a lack of adequate sanitation and waste removal facilities in the informal settlements, as well as poor management and disposal of sewage, the stormwater drainage pipes leading directly to the rivers are often used as a means of disposal of human and animal waste (SARDC, 2002).
The most common bacterial pollutants isolated from water sources include Escherichia coli (E. coli), Vibrio cholerae, Campylobacter, Salmonella, Shigella and Aeromonas hydrophila (Lightfoot, 2003). In addition, the occurrence of biofilms or encrustations in water sources, which harbour various types of micro-organisms (Banning et al., 2003), allows for the multiplication of pathogens such as Pseudomonas, Mycobacter, Campylobacter, Klebsiella, Aeromonas, Legionella spp., Helicobacter pylori and Salmonella typhimurium (Gião et al., 2008).
In South Africa, the presence of E. coli pathogens in sewage-contaminated river water and E. coli O157:H7 in sewage (Müller et al., 2001), was confirmed in an assessment conducted in 2003. Pathogens such as Salmonella, Shigella, Vibrio cholerae and coliphages have also been isolated from the final effluent of wastewater treatment facilities in the Eastern Cape, South Africa (Momba et al., 2006).
The aim of this study was to identify predominant bacterial species isolated from water and biofilm samples collected from 2 sites along the Berg and Plankenburg Rivers, in the Western Cape, South Africa. Population shifts between sites were also identified using 16S rRNA PCR and DNA sequencing. Phylogenetic analyses were performed by aligning grouped DNA sequences with the Infernal Secondary Structure Based Aligner within the Ribosomal Database Project (RDP) system.
Materials and methods
Sampling sites
A previous study identified 4 sampling sites along the Berg River (Paulse et al., 2007) and 4 sites along the Plankenburg River (Paulse et al., 2009). Based on the results obtained from these studies, deoxyribonucleic acid analysis was performed on samples (water and biofilm) collected from Sites A and B2 (Berg River) and from Sites A and B (Plankenburg River).
Sampling
One-litre water samples were collected in sterile Nalgenepolypropylene bottles (Cole-Palmer Instrument Company) at each sampling site, once a month, and stored on ice to maintain the lowest possible temperature. Materials such as stones, glass, leaves, etc., were also collected from each sampling site and stored in sterile whirlpack bags for subsequent biofilm isolation.
Sonication of biofilm samples
Glass, stones and leaves collected from the river were sonicated for 10 min in 60 ml sterile dH2O using a UMC 5 ultrasonication bath (Instrulab Inc.). The sonication step was repeated at least 3 times, depending on the amount of material collected from each sampling site. The bacterial suspension obtained was used for further microbiological analysis.
DNA extraction and agarose gel electrophoresis
Cultures from planktonic and sessile samples (sonicated bacterial suspension) obtained from the sampling sites were spreadplated onto Nutrient Agar (NA) (Merck, Biolab Diagnostics) after serial dilutions (10-1 to 10-7) were performed. Plates were incubated for 3-4 days at 37°C. Thereafter, distinct visible cells (colony forming units (CFU)) were identified based on morphological differences and re-streaked onto NA plates for isolation of pure cultures (approximately 824 isolates were selected for further analysis). Deoxyribonucleic acid (DNA) extraction was performed using the High Pure PCR Template Preparation Kit as per manufacturer's instructions (Roche Diagnostics). Extracted DNA samples (10 ul) were electrophoretically analysed on a 0.8% molecular grade agarose gel.
Polymerase chain reaction (PCR)
It should be noted that the PCR and DNA sequencing techniques were performed only on the culturable microorganisms isolated from the water and biofilm samples. The extracted DNA from individual samples was amplified using 2 primer sets, which amplify the 16S rRNA gene in bacteria. Amplification of target DNA samples (5 ul) (100 ng/50 ul) was performed in a total reaction volume of 50 ul containing a 10 mM dNTP mix (1 ul), 25 mM MgCl2 (4 ul), 5 x PCR Buffer with (NH4)2SO4 (10 ul), 10 uM forward (RW01) primer [AAC TGG AGG AAG GTG GGG AT] (2.5 ul), 10 uM reverse (DG74) primer [AGG AGG TGA TCC AAC CGC A] (2.5 ul) (Greisen et al., 1994), GoTaq DNA polymerase (0.25 ul) and sterile distilled H2O (24.75 ul). For the second primer set all of the reagents mentioned above were added proportionally, together with 10 uM forward (RDR080) primer [aAC TGG AGG AAG GTG GGG AC] (2.5 ul) and 10 uM reverse (DG74) primer [AGG AGG TGA TCC AAC CGC A] (2.5 ul) (Greisen et al., 1994) to obtain a total volume of 50 ul for subsequent amplification. The PCR procedure included an initial dena-turation step of 5 min at 95°C, followed by 30 cycles of amplification (25 s at 95°C, 25 s 55°C and 1 min at 72°C). The final extension step was performed at 72°C for 10 min.
16S ribosomal RNA sequencing
Successfully amplified PCR products (~600 bp) were purified using a High Pure PCR Product Purification Kit as per manufacturer's instructions (Roche Diagnostics). The DNA concentrations were determined using the QubitlTM fluorometer (Invitrogen) and the Quant-iTlTM dsDNA BR (Broad-range) Assay kit 2-1 000 ng as per manufacturer's instructions (Molecular probes and Invitrogen). The DNA samples (5-268 ng/ml = 0.005-0.3 ng/u£) were loaded onto 96-well plates (15 ui per sample), dried in a speed vac with medium heat for 30 to 60 min (depending on the volumes), and sent for subsequent sequencing. The Applied Biosystems Big Dye Terminator v3.1 Cycle sequencing Kit was used for the sequencing reactions, as per manufacturers' protocols. Sequences were identified using the Sequence Match Programme (Cole et al., 2009) obtained from the Ribosomal Database Project website.
Phylogenetic analysis
All of the DNA sequences obtained for the Berg and Plankenburg Rivers (Sites A and B) were grouped and aligned with the Infernal Secondary Structure Based Aligner in the Ribosomal Database Project (RDP). An unrooted tree was constructed using the Weighbour weighted neighbour-joining tree building algorithm within the RDP programme and viewed using the MEGA version 4.1 (Molecular Evolutionary Genetics Analysis 4.1) programme (Tamura et al., 2007). Branching patterns were evaluated by pairing 1 000 replicates of randomly-generated trees.
Results and discussion
Figure 1 (a and b) illustrates the amplified PCR products analysed on a 1.2% agarose gel, against a MassRulerlTM DNA Ladder Mix, #SM0403 (Fermentas), resembling the approximate size of 600 bp. The gel electrophoresis pictures indicating the representative samples (138 to 155) isolated at Site B in the Berg River were amplified using both Primer Sets 1 and 2 (Greisen et al., 1994). While most samples represented in Fig. 1a were successfully amplified with Primer Set 1, Samples 143, 147, 152 and 154 were not observed (Fig. 1b) when amplified with Primer Set 2. Samples sent for subsequent sequencing were therefore selected based on successful amplification with either primer set.
The phylogenetic trees depicted in Figs. 2 and 3 include various species of both Gram-positive and Gramnegative heterotrophic bacteria. According to Nikaido (1996), Gram-negative bacteria tend to be more resistant to various lipophilic and amphiphilic inhibitors than Gram-positive bacteria. This indicates that these organisms possess survival mechanisms which Gram-positive organisms lack and could explain why these organisms are surviving adverse environmental conditions. In all of the trees the Gram-negative group make up the largest number of organisms isolated and are further subdivided into the Enterobacteriaceae, Proteobacteria and the high G+C Gram-negatives.
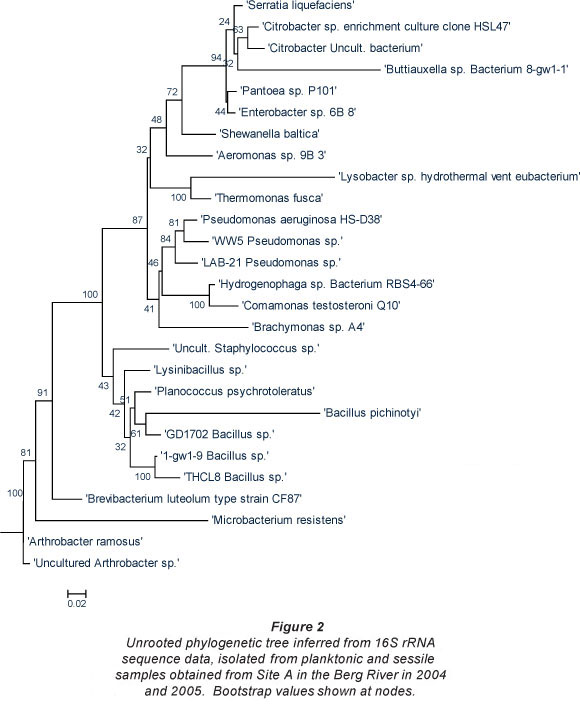
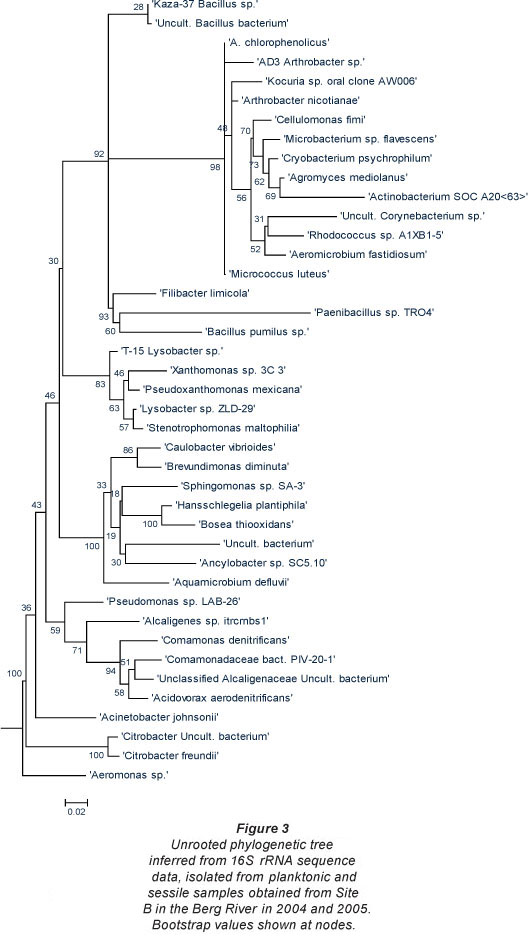
Berg River
Bacterial species isolated from the planktonic and sessile samples obtained from Site A (site situated upstream from the suspected point-source of pollution), are presented in Fig. 2. Phylogenetic analysis based on the 16S rRNA of the bacterial strains isolated in 2004 and 2005 displayed acceptable bootstrap values. In addition to the Enterobacteriaceae, the Proteobacteria and the high G+C Gram-negatives isolated from this site, the isolates from the low G+C Gram-positives were less diverse and consisted of Staphylococcus sp., various Bacillus spp. and the Firmicutes bacterium.
The Enterobacteriaceae, which are common and widespread in the environment, include the coliform bacteria, which are used as indicator organisms in evaluating faecal pollution in various water bodies (Figueras and Borrego, 2010). The fact that Enterobacter sp., Citrobacter sp., Serratia sp., Shewanella sp. and Aeromonas sp., amongst others, were detected at Site A is a clear indication of faecal contamination of the river in this area. These isolates belong to the indicator coliform group of organisms of which most are harmless. However, their presence in a water system could lead to major health concerns and may indicate the presence of potential disease-causing bacterial strains, i.e. pathogens (Figueras and Borrego, 2010). The presence of these faecal indicators in the Berg River also relates to a previous study (Paulse et al., 2007), where significantly (p < 0.05) high faecal coliform and E. coli counts were recorded by means of the most probable number technique (MPN).
In addition, the presence of Pseudomonas sp. and particularly, Pseudomonas aeruginosa, constitutes a potential health threat to children, the elderly and immune-compromised patients. This organism is an opportunistic pathogen, which, when sensing a breakdown in a host's defence system, takes advantage or exploits the situation thereby initiating infection (Todar, 2008). In addition, a study conducted by Miteva et al. (2004) indicated that isolates exhibiting multiple resistances, among the Proteobacteria, belong to the Pseudomonas spp. group.
The final group in this tree, the low G+C Gram-positives, consisted of Staphylococcus spp. and various Bacillus spp. Even though most Staphylococcus spp. are harmless and reside normally on the mucous membranes and skin of humans (Madigan and Martinko, 2005), these organisms may also cause various diseases in humans through either toxin production or penetration.
To add to the major health concerns of the inhabitants living in close proximity to this river, some of the bacterial species observed at Site A (i.e. agricultural area) were conserved when compared to microorganisms identified at Site B (Fig. 3). Hundreds of informal settlement dwellers as well as farm workers (agricultural area), live in close proximity to this site and many use the riverbanks as 'toilet schemes' as the sanitary facilities in the settlement are either in a poor state or completely non-existent in the vicinity.
As observed at Site A, the Gram-negative bacteria constituted the largest group of organisms isolated from this site. In addition to the Citrobacter sp. identified at this site and previously at Site A in the Berg River, bacterial species introduced into the river at Site B, included, amongst others, Micrococcus sp., Corynebacterium sp. and Rhodococcus sp. Even though most of these species are not well-known human pathogens, species such as Citrobacter freundii are opportunistic pathogens, which normally inhabit the intestine, and have been linked to alimentary infections and infections of the urinary tract, middle ear, gallbladder and meninges as well as bloodstream infections, brain abscesses, neonatal sepsis, intra-abdominal sepsis and pneumonia (Pepperell et al., 2002).
In addition to the Pseudomonas sp. previously mentioned, Stenotrophomonas maltophilia, was also isolated from Site B in the Berg River. Some of the organisms isolated at Site A remained dominant at Site B and included species such as Aeromonas sp., Comamonas sp., and Pseudomonas sp. In addition, Microbacterium, and various Bacillus spp., persisted at both Sites A and B.
Plankenburg River
Various members of the Enterobacteriaceae group were isolated from the Plankenburg River sampling sites thereby confirming faecal contamination in the river (phylogenetic trees not included). One of the isolated bacterial species was identified as the organism Yersinia enterocolitica subsp. (Kuhm, 2009), a disease which leads to self-limiting entero-colitis. Symptoms include watery or bloody diarrhoea and fever. The above-mentioned bacterial species, along with the Aeromonas sp., Alcaligenes faecalis and Acinetobacter sp., are all indicators of faecal contamination (Gerischer, 2008). The presence of these faecal indicators also confirms the results of a previous study (Paulse et al., 2009), where significantly (p<0.05) high faecal coliform and E. coli counts were recorded by means of the MPN technique. During this study, faecal coliform and E. coli counts of 3.5 x 106 microorganisms/100 ml were observed at Site B along the Plankenburg River.
Alcaligenes faecalis, which was originally isolated from faeces but has become more common in environmental samples, was isolated from Site A. Alcaligenes faecalis is normally found in soil, water and environments associated with humans. Even though the organism is generally regarded as non-pathogenic, it has previously been implicated as an opportunistic pathogen, which may lead to urinary tract infections (Meharwal et al., 2002).
Aeromonas sp., and particularly, A. hydrophila, is associated with gastroenteritis, cellulitis and other diseases in humans (Khajanchi et al., 2010). This species belongs to the Vibrionaceae which consist of Gram-negative, straight or curved rods with polar flagella. This family also includes the important Vibrio sp. of which V. cholerae and V. parahaemolyt-icus are the most well-known human pathogens. Vibrio chol-erae is responsible for the disease cholera, while V. parahaemo-lyticus infections result in gastroenteritis in humans after the ingestion of contaminated seafood (Tavana et al., 2010). Even though studies of rRNA homology suggest that Vibrio sp. and Aeromonas sp. differ more widely, previous studies have reported the difficulty in identifying Aeromonas to the geno-species level (Pavan et al., 2005; Janda and Abbot, 2010).
Apart from Y. enterocolitica isolated from Site B, other important bacterial species introduced at this site included various members of the Enterobacteriaceae group such as Citrobacter gilleni, C. braakii, Pantoea agglomerans and Enterobacter aerogenes, thereby again confirming faecal contamination at this site. As previously mentioned, this is the point of the river that is closest to the informal settlement. It is also the site where faecal wastewater from nearby sanitary facilities is flushed into the river. Bacterial species from the genus Yersinia are considered major human pathogens and may be the causative agent of plague in susceptible individuals. Overall, the species diversity of micro-organisms isolated from Site B was lower in comparison to Site A.
Various species of the Gram-positive Bacillus genus were also identified at both sites in the Plankenburg River. Bacillus species isolated included various unidentified species as well as B. aquimaris, B. thuringiensis, and B. pumilus. In addition, Bacillus cereus, which may cause food poisoning in humans, was also isolated and identified. Bacillus cereus infections may lead to severe nausea, vomiting and diarrhoea (Lampel et al., 2012). In general, Bacillus spp. can occupy a wide variety of ecological niches and its spores are found ubiquitously in the environment (Pillai et al., 2006).
Conclusions
The major conclusions of the study are as follows:
Phylogenetic analyses showed that Gram-negative microorganisms dominated at all of the sites sampled in both the Berg and Plankenburg River systems;
Bacterial species from the Enterobacteriaceae family were isolated from all sampling sites in both river systems. This indicates that both the Berg and Plankenburg Rivers were subjected to faecal pollutants.
A similar trend in population shift was observed between the sampled sites for both river systems:
- In certain instances, isolates observed at Site A (i.e.
before the point sources of pollution) did not persist in the river systems and were thus not identified at Site B (i.e. after the point sources of pollution), e.g. Serratia liquefaciens, isolated from Site A in the Berg River, was not detected at Site B in the river system.
- Similarly, various species (e.g. Yersinia enterocolitica subsp. Enterocolitica, at Site B in Plankenburg River) were also introduced at Site B, which were not identified at Site A; this could be ascribed to the introduction of microbial pollutants from the surrounding environment as follows:
- Stormwater drainage pipes from the adjacent informal settlements, enter both river systems at Site B.
° Along the Plankenburg River, Site B is also located close to an industrial area.
° Site B along the Berg River is in close proximity to an agricultural area which could also contribute to the pollutant load.
The predominant pathogenic microorganisms isolated from the Berg River included: at Site A, Pseudomonas aeruginosa, Comamonas testosteroni, and Aeromonas sp.; at Site B, Bacillus pumilus, Stenotrophomonas maltophilia, Sphingomonas sp. and Aeromonas sp. Various opportunistic pathogens, including Serratia liquefaciens, Micrococcus sp. and Cellulomonas sp. were also isolated from the Berg River.
In the Plankenburg River, pathogens isolated included Pseudomonas sp., Yersinia enterocolitica, Bacillus pumilus, Bacillus cereus and Citrobacter braaki (Site A); B. pumilus, Pseudomonas sp., Comamonas testosteroni and Aeromonas sp. were isolated from Site B.
Acknowledgements
The National Research Foundation (NRF) and Cape Peninsula University of Technology (CPUT) are thanked for financial support.
References
BANNING N, TOZE S and MEE BJ (2003) Persistence of biofilm-associated Escherichia coli and Pseudomonas aeruginosa in groundwater and treated effluent in laboratory model system. Microbiology 149 47-55. [ Links ]
COLE JR, WANG Q, CARDENAS E, FISH J, CHAI B, FARRIS RJ, KULAM-SYED-MOHIDEEN AS, McGARRELL DM, MARSH T, GARRITY GM and TIEDJE JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37 D141-D145. [ Links ]
FIGUERAS MJ and BORREGO JJ (2010) New perspectives in monitoring drinking water microbial quality. Int. J. Environ. Res. Public Health 7 4179-4202. [ Links ]
GERISCHER U (ed.) (2008) Acinetobacter Molecular Biology (1st edn.). Caister Academic Press, Germany. [ Links ]
GIÃO MS, AZEVEDO NF, WILKS SA, VIEIRA MJ and KEEVIL CW (2008) Persistence of Helicobacter pylori in heterotrophic drinking-water biofilms. Appl. Environ. Microbiol. 74 (19) 5898-5904. [ Links ]
GREISEN K, LOEFFELHOLZ M, PUROHIT A and LEONG D (1994) PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32 335-351. [ Links ]
JANDA JM and ABBOTT SL (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23 (1) 35-73. [ Links ]
KHAJANCHI BK, FADL AA, BORCHARDT MA, BERG RL, HORNEMAN AJ, STEMPER ME, JOSEPH SW, MOYER NP, SHA J and CHPRA AK (2010) Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission. Appl. Environ. Microbiol. 76 (7) 2313-2325. [ Links ]
KUHM AE, SUTER D, FELLEISEN R and RAU J (2009) Identification of Yersinia enterolitica at the species and subspecies levels by Fourier Transform Infrared Spectroscopy. Appl. Environ. Microbiol. 75 (18) 5809-5813. [ Links ]
LAMPEL KA, SUFIAN AL-KHALDI S and CAHILL SM (2012) Bad Bug Book l Handbook of Foodborne Pathogenic Microorganisms and Natural Toxins (2nd edn.). United States Food & Drug Association, Giza. [ Links ]
LIGHTFOOT NF (2003) Bacteria of potential health concern. In: Bartram K, Cotruvo J, Exner M, Fricker C and Glasmacher A (eds.) Heterotrophic Plate Counts and Drinking-water Safety. IWA Publishing, UK. [ Links ]
MADIGAN M and MARTINKO J (editors) (2005) Brock Biology of Microorganisms (11th edn.). Prentice Hall, Upper Saddle River. [ Links ]
MEHARWAL SK, TANEJA N, SHARMA SK and SHARMA M (2002) Complicated nosocomial UTI caused by nonfermentors. Indian J. Urol. 18 123-128. [ Links ]
MITEVA VI, SHERIDAN PP and BRENCHLEY JE (2004) Phylogenetic and physiological diversity of micro-organisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 70 202-213. [ Links ]
MOMBA MNB, KFIR R, VENTER SN and CLOETE TE (2006) An overview of biofilm formation in distribution systems and its impact on the deterioration of water quality. Water SA 26 (1) 59-66. [ Links ]
MULLER EE, EHLERS MM and GRABOW WOK (2001) The Occurrence of E. coli 0157:H7 in South African Water Sources intended for direct and indirect human consumption. Water Res. 35 3085-3088. [ Links ]
NIKAIDO H (1996) Multidrug Efflux Pumps of Gram-Negative Bacteria. J. Bacteriol. 178 (20) 5853-5859. [ Links ]
PAULSE AN, JACKSON VA and KHAN W (2007) Comparison of enumeration techniques for the investigation of bacterial pollution in the Berg River, Western Cape, South Africa. Water SA 33 (2) 165-173. [ Links ]
PAULSE AN, JACKSON VA and KHAN W (2009) Comparison of microbial contamination at various sites along the Plankenburg-and Diep Rivers, Western Cape, South Africa. Water SA 35 (4) 469-478. [ Links ]
PAVAN ME, FRANCO RJ, RODRIGUEZ JM, GADALETA P, ABBOTT SL, JANDA JM and ZORZÖPULOS J (2005) Phylogenetic relationships of the genus Kluyvera: transfer of Enterobacter intermedins Izard et al. 1980 to the genus Kluyvera as Kluyvera intermedia comb. nov. and reclassification of Kluyvera cochleae as a later synonym of K. intermedia. Intern. J. System. Evol. Microbiol. 55 437-442. [ Links ]
PEPPERELL C, KUS JV, GARDAM MA, HUMAR A and BURROWS LL (2002) Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 46 3555-3560. [ Links ]
PILLAI A, THOMAS S and ARORA J (2006) Bacillus cereus: The forgotten pathogen. Surg. Infect. 7 (3) 305-308. [ Links ]
SOUTHERN AFRICAN RESEARCH AND DOCUMENTATION CENTRE (2002) Defining and mainstreaming environmental sustainability in water resources management in Southern Africa. URL: http://databases.sardc.net/books/MainWB/index.php (Accessed 7 May 2008). [ Links ]
TAMURA K, DUDLEY J, NEI M and KUMAR S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.1. Mol. Biol. Evol. 24 1596-1599. [ Links ]
TAVANA AM, FALLAH Z, ZAHRAEE SM, ASL HM, RAHBAR M, MAFI M and ESMI N (2010) Effects of climate on the cholera outbreak in Iran during seven years (2000-2006). Ann. Trop. Med. Public Health 1 (2) 43-46. [ Links ]
TODAR K (2008) Todar's Online Textbook of Bacteriology. URL: http://textbookofbacteriology.net/pseudomonas.html (Accessed 6 October 2011). [ Links ]
Received 27 October 2009; accepted in revised form 2 October 2012.
* To whom all correspondence should be addressed. ffi +27 21 808 5804; fax: +27 21 808 5846; e-mail: wesaal@sun.ac.za














