Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Water SA
On-line version ISSN 1816-7950
Print version ISSN 0378-4738
Water SA vol.37 n.1 Pretoria Jan. 2011
Biodegradation characterisation and kinetics of m-cresol by Lysinibacillus cresolivorans
Haiyan YaoI; Yuan RenI,II,III,*; Chaohai WeiI,II,III; Siyang YueI
ICollege of Environmental Science and Engineering, South China University of Technology, China
IIThe Key Laboratory of Pollution Control and Ecosystem Restoration in Industry Clusters, Ministry of Education, China
IIIThe Key Laboratory of Environmental Protection and Eco-Remediation of Guangdong Regular Higher Education Institutions, China
ABSTRACT
A novel strain of m-cresol-degrading bacterium, named as Lysinibacillus cresolivorans, was isolated from aerobic sludge from a coking wastewater treatment plant. This bacterium is able to utilise m-cresol as its sole source of carbon and energy. The optimal pH for growth is 6.8 ~ 7.3 and the optimal temperature is 35ºC. Compared to organic nitrogen sources, inorganic nitrogen sources were easily utilised for the m-cresol biodegradation. The degradation rate of m-cresol at different starting concentrations was analysed with zero-order kinetic characteristics. When the initial concentration of m-cresol was 224.2 mg·ℓ-1, the reaction rate reached a maximum at 46.80 mg·(ℓ·h)-1.The cell growth kinetics was also investigated with initial m-cresol concentrations varying from 0 to 1 200 mg·ℓ-1. The growth kinetics was well described by the Haldane kinetic models. The parameter values of m-cresol on cell growth were µmax = 0.89 h-1, Ks = 426.25 mg·ℓ-1, Ki = 51.26 mg·ℓ-1. Experiments supplementing growth with glucose indicated that this substrate increased the biomass, and also induced the biodegradation of m-cresol. From the results, it can be concluded that Lysinibacillus cresolivorans is an efficient m-cresoldegrading bacterium and that glucose plays multiple roles in the co-substrate condition.
Keywords: m-cresol; biodegradation; Lysinibacillus cresolivorans; high efficient degrading bacterium
Introduction
Phenolic compounds are environmental pollutants because of their wide distribution and detection in the effluent of various industrial operations, which include petrochemicals, textile, dying, varnish industries, phenolic resin manufacturing and steel plants (Jiang et al., 2006; Juang and Tsai, 2006). These compounds have high toxicity to most of the microorganisms, plants, fish and animals, which can cause considerable damage to the environment. The aromatic ring structure in these compounds isrelatively resistant to natural biodegradation (Juang and Tsai, 2006). Series of available processes have been applied to eliminate these recalcitrant chemicals before discharge into water bodies. Studies have demonstrated that biological treatment shows more promise for wastewater treatment of phenolic compounds (Bai et al., 2007) than conventional physicochemical treatment techniques such as adsorption, coagulation, ion exchange, chemical oxidation, photocatalytic oxidation and other advanced oxidation processes.
Previous studies have reported that phenolic compounds can be degraded by microorganisms, most commonly Pseudomonas (Taylor, 1975; Ahamad et al., 2004; Goswami et al., 2005; Yang and Lee, 2007; Saravanan et al., 2008a). Pseudomonas can degrade phenol at less than 1 200 mg·ℓ-1 (Ren et al., 2009). m-Cresol is a representative phenolic compound and is listed as a priority pollutant by the United States Environmental Protection Agency (Bai et al., 2007). There are several reports of m-cresol biodegradation by Alcaligenes faecalis, Pseudomonas sp., and Candida albicans (Bai et al.,2007; Saravanan et al., 2008a; Wang et al., 2009). Ahamad and Kunhi (1999) reported that Pseudomonas sp. CP4 can mineralise all 3 isomers of cresol and the degradation rates of these 3 isomers were in the order: ortho- > para- > metacresol. Lag periods were observed with all 3 isomers, the longest being in the case of m-cresol because of the inactivation of the meta- methyl group. Considerable progress has been made, but most of the bacteria studied can only degrade m-cresol at concentrations of less than 600 mg·ℓ-1. In this paper, a novel bacterium capable of biodegrading m-cresol was isolated from the aerobic sludge from an efficient coking wastewater treatment plant. Morphological and biochemical analyses, including 16S rRNA gene sequence comparisons, demonstrated that the selected strain belongs to a novel species, Lysinibacillus cresolivorans. In order to fully characterise this new bacterium, its degradation characteristics and kinetics were examined.
Experimental
Chemicals and culture medium
m-Cresol (99.5% purity) was purchased from Sinopharm Chemical Reagent CO., Ltd, China. Other chemicals used in this study were all of analytical grade.
The mineral salts medium (MSM) used to culture the strain was: K2HPO4 2.24 g·ℓ-1, KH2PO4 2.75 g·ℓ-1, (NH4)SO4 1.00 g·ℓ-1, MgCl2·6H2O 0.2 g·ℓ-1, NaCl 0.1 g·ℓ-1, FeCl3·6H2O 0.02 g·ℓ-1, CaCl2 0.01 g·ℓ-1. m-Cresol was filtered, sterilised and added after autoclaving.
LB culture medium used to culture the strain was: beef extract 3.0 g·ℓ-1, peptone 10.0 g·ℓ-1, NaCl 5.0 g·ℓ-1, pH 7.0-7.2.
Enrichment and isolation of m-cresol-degrading strains
Activated sludge obtained from the aerobic basin of a coking wastewater plant at Shaoguan City was inoculated into 100 mℓ of a fresh sterile MSM containing 50 mg·ℓ-1 m-cresol as the sole carbon and energy source. Cultures were incubated at 35ºC at 170 r·min-1 and sampled periodically for m-cresol loss and turbidity due to bacterial growth. When the culture medium became turbid, which indicated growth, 10 mℓ of the suspension was transferred to a fresh sterile MSM containing 100 mg·ℓ-1 m-cresol. The procedure was repeated 4 times, to increase the level of m-cresol to 500 mg·ℓ-1. The final enriched culture was spread on MSM agar plates with 200 mg·ℓ-1 m-cresol as sole carbon source. After incubation for 24 h, different colonies were selected and isolated by using the streak plating method 3 times on LB plates. Finally, the purified strain was obtained. The strain was inoculated into a LB culture medium, incubated in a rotary shaker for 48 h at 35ºC, 170 r·min-1, and used for further degradation experiments.
DNA extraction and PCR amplification
For 16S rRNA gene sequencing, DNA was extracted and the almost complete 16S rRNA gene was amplified using primers F27 (5'-AGA GTT TGA TCC TGG CTC AG-3') and R1522 (5'-AAG GAG GTG ATC CAG CCG CA-3'). All reactions were carried out in a 50 µℓ volume, containing 36.5 µℓ deionized H2O , 5.0 µℓ 10×PCR buffer, 5.0 µℓ 2.5 mM dNTPs, 1.0 µℓ 10 µM primer F27, 1.0 µℓ 10 µM primer R1522, 1.0 µℓ DNA template, and 0.5 µℓ 5U(µℓ) -1 Blend-Taq. PCR amplification was conducted in an automated thermal cycler (PCR 9600, PE, USA) using the following protocol: initial denaturation for 3 min at 94ºC and 30 cycles of denaturation for 1 min at 94ºC, annealing for 1 min at 52ºC, extension for 1 min at 72ºC, followed by a final extension for 7 min at 72ºC. The 1.5kb PCR fragments were sequenced at the Shanghai Bioasia (Invitrogen) Biotechnology Co. Ltd. and the sequences were logged in Genbank to compare with registered/submitted sequences.
Biodegradation of m-cresol by Lysinibacillus cresolivorans
The strain was inoculated into sterile MSM supplemented with m-cresol as the sole carbon and energy source, and all cultures were incubated in a rotary shaker at 170 r·min-1. This was used for studying the optimisation of growth parameters: temperature, pH, inoculation dosage, nitrogen, and carbon sources. When optimal growth conditions were set, m-cresol concentrations varying from 54.1 to 529.1 mg·ℓ-1 were investigated. Each treatment was conducted in triplicate and the mean values obtained are reported. The degradation efficiency was determined and estimated by the remaining rate of m-cresol degradation from the liquid culture.
Analytical method
Cell density in the culture medium was estimated with a UV-2800 spectrophotometer (Unico, Shanghai, China) by measuring absorbance (OD) at 600 nm. The quantification of m-cresol was measured by high performance liquid chromatography (HPLC). HPLC was performed with C18 column (Agilent HC-C18, 5 µm, 4.6 × 250 mm) with methanol/water (60/40, v/v) as the mobile phase at a flow rate of 0.7 mℓ/min, and detection was realised with a UV detector at 270 nm. The retention time for m-cresol was 6.67 min.
Results and discussion
Isolation and identification of Lysinibacillus cresolivorans
After multiple cycles of inoculation and screening, the colony capable of degrading m-cresol was obtained from the enrichment culture. The nearly complete 16S rRNA gene of the purified isolate was PCR-amplified and sequenced. BLAST search of its 16S rRNA gene sequence indicates that the strain shares 94% identity with Lysinibacillus fusiformis and Lysinibacillus macroide, which indicated that it belongs to a novel Lysinibacillus sp. strain. (The PCR, sequencing and physiological/biochemical identification results have been submitted to IJSEM). Its GenBank accession number is EU043375 and the collection number of Lysinibacillus cresolivorans is CCTCC M 208210 T and NRRL B-59352T.
Growth on m-cresol
Lysinibacillus cresolivorans suspension was inoculated into MSM (10%, v/v) containing 52.76 mg·ℓ-1 m-cresol at exponential phase (OD = 0.083). The culture was then incubated in a rotary shaker at 35ºC, 170 r·min-1. Cell density and residual concentration of m-cresol were estimated at the same time points. The growth of Lysinibacillus cresolivorans on m-cresol is shown in Fig. 1. We can see that Lysinibacillus cresolivorans utilised m-cresol as its sole source of carbon and energy, and growth of bacteria is closely related to the biodegradation of m-cresol.
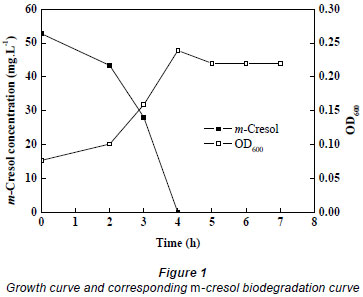
Degradation of m-cresol by Lysinibacillus cresolivorans
The hydrogen ion concentration in the culture medium influences bacterial growth since pH limits the activity of enzymes (Wu et al., 2009). The effect of pH on m-cresol biodegradation by Lysinibacillus cresolivorans is shown in Fig. 2. The highest biodegradation rate was achieved when the pH ranged from 6.9 to7.3. This indicates that a neutral environment is suitable for biodegradation of m-cresol. When the pH was less than 5.0 or greater than 9.0, the degradation rate of m-cresol was decreased. A long lag phase was observed when the pH was less than 4.9, with more than 80% residual m-cresol remaining after 6 h biodegradation.
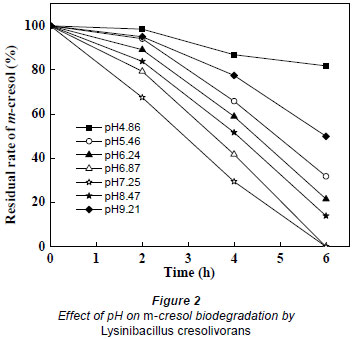
Bai et al. (2007) isolated Alcaligenes faecalis, capable of degrading m-cresol in media in which the pH was higher than 9.0 or lower than 5.0. The highest biodegradation rate was achieved at pH 7.0. The optimal pH value for m-cresol biodegradation by Lysinibacillus cresolivorans was 7.0 in this study.
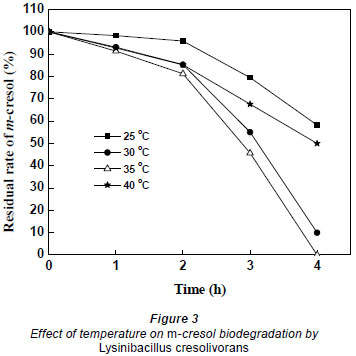
In order to optimise biodegradation, the temperature for m-cresol biodegradation was set at 25, 30, 35, and 40ºC; the starting concentration of m-cresol was aprox. 50 mg·ℓ-1. As shown in Fig. 3, when the temperature was 35ºC m-cresol was completely degraded after 4 h incubation; however, the residual percentage was around 58%, 10% and 50% at 25, 30 and 40ºC, respectively. Therefore, the optimal temperature, at which the degradative enzyme reached highest activity, was 35ºC.
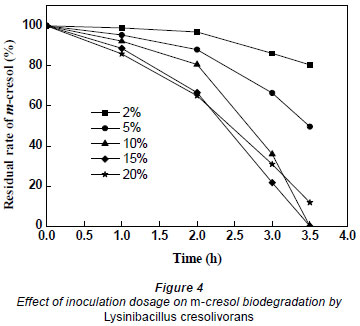
In order to study the effects of inoculation dosage on m-cresol biodegradation, the inoculation dosage of bacteria was changed from 2% to 20%, with initial optical density of culture at 0.032, 0.056, 0.083, 0.105 and 0.137. As shown in Fig. 4, the biodegradation rate increased as inoculation dosage was increased from 2% to 15%. m-Cresol had been completely degraded after 3.5 h when the inoculation dosage was 10% and 15%. However, at the same time, the removal percentages of m-cresol were 19.5%, 50.3% and 88.2% at 2%, 5%, and 20% dosage, respectively. When inoculation dosage was up to 20% or more, the biodegradation decreased. Surplus microorganisms competed for the reduced amount of nutrition which led to a declining biodegradation rate. Therefore, 10% was found to be the optimal inoculation dosage for m-cresol biodegradation from economical and efficient consideration.
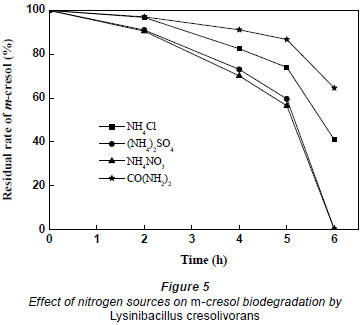
Nitrogen is important for growth and metabolism. NH4Cl, (NH4)2SO4, NH4NO3 and CO(NH2)2 were compared in the biodegradation of m-cresol. Nitrogen content in every culture medium was around 100 mg·ℓ-1. The effect of nitrogen source on m-cresol biodegradation by Lysinibacillus cresolivorans is shown in Fig. 5. Inorganic nitrogen had a greater effect on the biodegradation of m-cresol than organic nitrogen (urea). (NH4)2SO4 and NH4NO3 have the best efficiency, at 6 h, for m-cresol biodegradation.
Biodegradation kinetics of m-cresol by Lysinibacillus cresolivorans
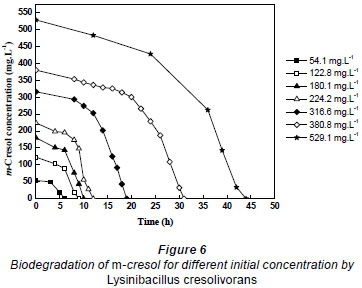
Batch cultures of Lysinibacillus cresolivorans were inoculated in MSM containing m-cresol as the sole carbon and energy source, in order to obtain the biodegradation kinetic model parameters of m-cresol. Substrate concentrations were measured over time for different initial concentrations of m-cresol (54.1 to 529.1 mg·ℓ-1) at optimal biodegradation conditions. Figure 6 shows that m-cresol at concentrations of 54.1, 122.8, 180.1, 224.2, 316.6, 380.8 and 529.1 mg·ℓ-1 was completely biodegraded by the culture in 6, 9, 10, 12, 19, 31 and 44 h, respectively. The lag period in each case was, respectively, 2, 4, 6, 8, 12, 21and 30 h. The results clearly showed good ability in biodegrading m-cresol, even at high concentrations, when employing Lysinibacillus cresolivorans.
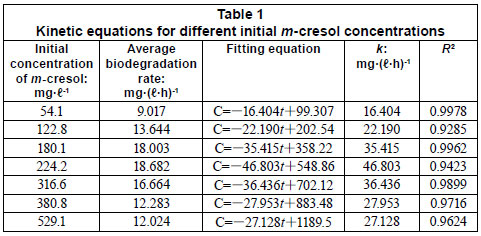
The average biodegradation rate and kinetic model equation of different m-cresol concentrations is represented in Table 1. The biodegradation of m-cresol was consistent with the zero-order kinetics equation when the lag period was excluded. The average biodegradation rate increased with an increase in initial m-cresol concentration, but the average biodegradation rate reached its maximum of 18.682 mg·(ℓ·h)-1 when m-cresol was 224.2 mg·ℓ-1. As the concentration continued to increase, the average biodegradation rate decreased from 18.68 mg·(ℓ·h)-1 to 12.02 mg·(ℓ·h)-1.
The biodegradation kinetics of m-cresol has been reported in prior studies. Saravanan et al. (2008b) investigated biodegradation kinetics of m-cresol by an indigenous mixed microbial culture isolated from a sewage treatment plant.
The substrate m-cresol was completely utilised within 21 h when present at concentrations of 100-600 mg·ℓ-1; a maximum time of 187 h was observed for its removal. Therefore, compared with other bacteria reported to biodegrade m-cresol, Lysinibacillus cresolivorans displays superior performance.
Growth kinetics of Lysinibacillus cresolivorans
Biodegradation experiments of Lysinibacillus cresolivorans were conducted with m-cresol concentrations ranging from 0 to 1 200 mg·ℓ-1. The experimental data were utilised for calculating the specific growth rate and culture biomass yield according to the following equations:

where
µx is the specific growth rate (h-1)
γx the cell growth rate
Cx the cell concentration (mg·ℓ-1).
Because of the substrate inhibition on the cell growth, Haldane's equation was selected due to its wide applicability for assessing the growth kinetics of inhibitory substrate (Haldane, 1965):

where:
S is the substrate concentration (mg·ℓ-1),
µmax is the maximum growth rate (h-1),
Ks is the substrate saturation coefficient and
Ki is the substrate inhibition coefficient.
The values of the parameters: µmax= 0.89 h-1, Ks = 426.25 mg·ℓ-1, Ki = 51.26 mg·ℓ-1, were derived using a nonlinear leastsquares regression analysis of ORIGIN 7.5 based on the experimental data obtained in the tests. The value of the squared 2-norm of the residual at these parameters was very small (3.0 × 10-4), which indicated that the regression curve agreed with the data very well.
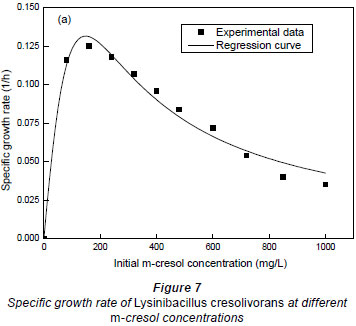
It could be seen in Fig. 7 that the maximum specific growth rate occurred at very low m-cresol concentrations. When the initial concentration of m-cresol was 151.5 mg·ℓ-1, the maximum specific growth rates were 0.1316 h-1. With the increase of initial substrate concentration, the specific growth rate decreased, which may be a result of an intense substrate inhibition. A higher substrate concentration demonstrated a stronger substrate inhibitory response.
The effect of glucose on m-cresol biodegradation
When the initial concentrations of m-cresol were, respectively, 380.8 and 529.1 mg·ℓ-1, it took 31 h and 43 h for complete biodegradation; the average biodegradation rate decreased to 12.283 and 12.024 mg·(ℓ·h)-1. Thus, heavy concentrations of substrate inhibited the biodegradation of m-cresol; the higher the m-cresol concentration, the stronger the substrate inhibition. In order to mitigate the inhibition, different concentrations of glucose were added to the culture containing 529.1 mg·ℓ-1 m-cresol. The glucose concentration changed from 0 to 800 mg·ℓ-1. As shown in Fig. 8, glucose promoted the biodegradation of m-cresol. Biodegradation time became shorter when glucose varied from 0 to 600 mg·ℓ-1, but when glucose concentration was further increased, biodegradation time increased. Therefore, the higher dosage of glucose was not better for the degradation of m-cresol. It took 22 h to completely biodegrade m-cresol at a glucose concentration of 600 mg·ℓ-1, while the removal percentage was only 17.61% without glucose at the same incubation duration.

Glucose is an easily degradable organic substance that is apt to be utilised by microbes. The mechanism by which glucose enhances the biodegradation of m-cresol may rely on increase in biomass only or on inducing the biodegradation of m-cresol, or both.. In this study, the OD of cultures when 600 mg·ℓ-1 glucose is provided as sole carbon source is 0.626. The biodegradation efficiencies of adding 600 mg·ℓ-1 glucose and adding 0.626 OD biomass were compared. If the 2 rates are the same, it can be concluded that glucose merely acts as an extra carbon source; if the former rate is higher, it can be concluded that glucose has an induction effect on m-cresol biodegradation.
We designed 4 experiments to study the effect of 600 mg·ℓ-1 glucose on the degradation of m-cresol according to the above hypothesis:
• Culture 1: 5 mℓ of inocula + 529 mg·ℓ-1 m-cresol
• Culture 2: 5 mℓ of inocula + 529 mg·ℓ-1 m-cresol + 600 mg·ℓ-1 glucose (at the same time)
• Culture 3: 5 mℓ of inocula + 600 mg·ℓ-1 glucose (at first); when OD600 reached 0.626, 529 mg·ℓ-1 m-cresol was also added
• Culture 4: 5 mℓ inocula + 529 mg·ℓ-1 m-cresol + additional biomass (OD=0.626)
All of the cultures were in 50 mℓ MSM media. Residual concentration of m-cresol was estimated at certain times. It could be concluded that the glucose and m-cresol were degrading simultaneously in Culture 2, as there were some intermediate products of glucose degradation. Culture 3 had little intermediate products of glucose degradation. The experiments were designed to confirm the presence of an induction effect. Culture 4 was used to estimate the effect of more dosage. All of the cultures can be compared with Culture 1 to illustrate the effect of glucose on degradation of m-cresol.
The results of 4 control experiments are shown in Fig. 9. Adding glucose and increasing the dosage can increase the degradation of m-cresol. As explained above, if glucose only has the effect of proliferation, Cultures 2 and 4 should have the same degrading efficiency. However, it was found that Culture 2 took less time than Culture 4, which indicates that glucose does not only play a role by increasing the biomass. In Culture 3, glucose was not completely utilised by microorganisms until the exponential phase. When m-cresol was added, there was little glucose to induce the biodegradation of m-cresol. Because the carbon source (glucose) had been exhausted, the activity of the microorganisms had decreased. Thus the efficiency of degradation was weaker for Culture 3 than Culture 4.
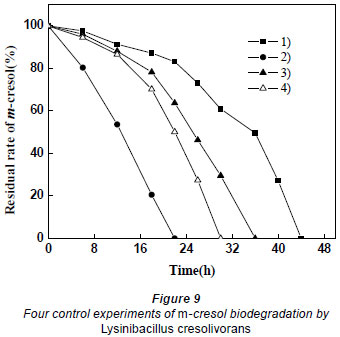
Based on the above results, it can be concluded that glucose not only increased the amount of bacteria, but also induced the biodegradation of m-cresol and shortened the lag phase. A simple increase of biomass cannot attain the equivalent degradation effect obtained when glucose was added to the culture. In the presence of an external carbon source, Lysinibacillus cresolivorans was still able to utilise m-cresol. It is thus favourable for biodegrading phenolic sewage containing other carbon pollutants.
Conclusions
Lysinibacillus cresolivorans, capable of using m-cresol as the sole source of carbon and energy, was isolated from the activated sludge in a coking wastewater treatment plant. It was identified by morphological and biochemical analyses, as well as 16S rRNA gene sequence comparisons, which demonstrated that the selected strain belongs to a novel species, Lysinibacillus cresolivorans. The optimal pH and temperature of m-cresol biodegradation by Bacillus cresolum was around 7.0 and 35ºC, respectively, and the most suitable inoculation dosage was 10%. The presence of a nitrogen source improved the biodegradation rate, NH4Cl, (NH4)2SO4 and NH4NO3 were better than CO(NH2)2 for the biodegradation of m-cresol. Under optimal growth conditions, the biodegradation of m-cresol changed from 54.1 to 529.1 mg·ℓ-1, and followed a zero-order kinetics equation. The maximum average biodegradation rate was 18.682 mg·(ℓ·h)-1 when the initial m-cresol concentration was 224.2 mg·ℓ-1. The experimental data of growth kinetics of Lysinibacillus cresolivorans fitted the Haldane kinetics models well. The parameter values of m-cresol on cell growth were µ max = 0.89 h-1, Ks = 426.25 mg·ℓ-1, Ki = 51.26 mg·ℓ-1. Glucose accelerated the biodegradation rate, especially in the cultures containing a high concentration of m-cresol. When glucose concentration was 600 and 800 mg·ℓ-1, respectively, complete biodegradation of 529.1 mg·ℓ-1 m-cresol only took 22 and 24 h, compared to 43 h without glucose; biodegradation time was thus halved. Four control experiments showed that glucose had an induction effect on the biodegradation of m-cresol. Lysinibacillus cresolivorans may prove to be an ideal microorganism for eliminating phenolic compounds.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 20977035), Research Fund of The Guangdong Provincial Laboratory of Pollution Control and Ecological Restoration and The Key Laboratory of Pollution Control and Ecosystem Restoration in Industry Clusters, Ministry of Education, China. The authors thank Dr Jessica McCormick for her help with linguistic revision.
References
AHAMAD PYA and KUNHI AAM (1999) Degradation of high concentrations of cresols by Pseudomonas sp. CP4. World J. Microbiol. Biotechnol. 15 (2) 321-323. [ Links ]
AHAMAD PYA, KUNHI AAM and DIVAKAR S (2004) New metabolic pathway for o-cresol degradation by Pseudomonas sp. CP4 as evidenced by 1H NMR spectroscopic studies. World J. Microbiol. Biotechnol. 17 (4) 371-377. [ Links ]
BAI J, WEN JP, LI HM and JIANG Y (2007) Kinetic modeling of growth and biodegradation of phenol and m-cresol using Alcaligenes faecalis. Process Biochem. 42 (4) 510-517. [ Links ]
GOSWAMI M, SHIVARAMAN N and SINGH RP (2005) Microbial metabolism of 2-chlorphenol, phenol and p-cresol by Rhodococcus erythropolis M1 in co-culture with Pseudomonas fluorescens P1. Microbiol. Res. 160 101-109. [ Links ]
JIANG Y, WEN JP, BAI J, WANG DQ and HU ZD (2006) Phenol biodegradation by the yeast Candida tropicalis in the presence of m-cresol. Biochem. Eng. J. 29 (3) 227-234. [ Links ]
JUANG RS and TSAI SY (2006) Growth kinetics of Pseudomonas putida in the biodegradation of single and mixed phenol and sodium salicylate. Biochem. Eng. J. 31 (2) 133-140. [ Links ]
REN Y, PENG LH, DENG LJ and WEI CH (2009) Isolation and Characterization of Citrobacter farmeri SCO1: A Novel m-Cresol-Degrading Strain. Environ. Eng. Sci. 26 (10) 1489-1495. [ Links ]
SARAVANAN P, PAKSHIRAJAN K and SAHA P (2008a) Biodegradation of phenol and m-cresol in a batch and fed batch operated internal loop airlift bioreactor by indigenous mixed microbial culture predominantly Pseudomonas sp. Bioresour. Technol. 99 (18) 8553-8558. [ Links ]
SARAVANAN P, PAKSHIRAJAN K and SAHA P (2008b) Kinetics of phenol and m-cresol biodegradation by an indigenous mixed microbial culture isolated from a sewage treatment plant. J. Environ. Sci. CHINA 20 (12) 1508-1513. [ Links ]
TAYLOR DJHA (1975) Pathways for the degradation of m-cresol and p-cresol by Pseudomonas putida. J. Bacteriol. 122 (1) 1-6. [ Links ]
WANG GY, WEN JP, LI HM and QIU CS (2009) Biodegradation of phenol and m-cresol by Candida albicans PDY-07 under anaerobic condition. J. Ind. Microbiol. Biotechnol. 36 (6) 809-814. [ Links ]
WU HZ, WEI CH, WANG YQ, HE QC and LIANG SZ (2009) Degradation of o-chloronitrobenzene as the sole carbon and nitrogen sources by Pseudomonas putida. OCNB-1 J. Environ. Sci. CHINA 21 (1) 89-95. [ Links ]
YANG CF and LEE CM (2007) Enrichment isolation, and characterization of phenol-degrading Pseudomonas resinovorans strain P-1 and Brevibacillus sp. strain P-6. Int. Biodeterior. Biodegrad. 59 (3) 206-210. [ Links ]
Received 22 April 2010; accepted in revised form 8 November 2010.
* To whom all correspondence should be addressed. +86-20-39380588; fax: +86-20-39380588; e-mail: ceyren@scut.edu.cn














