Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
Water SA
versão On-line ISSN 1816-7950
versão impressa ISSN 0378-4738
Water SA vol.36 no.2 Pretoria Jan. 2010
The occurrence of pathogenic Escherichia coli in South African wastewater treatment plants as detected by multiplex PCR
KB Omar*; TG Barnard
Water and Health Research Unit, Faculty of Health Sciences, University of Johannesburg, PO Box 17011, Doornfontein 2028, Johannesburg, South Africa
ABSTRACT
The aim of this study was to investigate the use of PCR to detect commensal and diarrhoeagenic Escherichia coli concentrated from water samples using membrane filtration. To achieve this, culture-based and PCR-based methods were compared for the detection of E. coli in raw sewage and primary, secondary and tertiary effluents from 6 wastewater treatment plants around Johannesburg, Gauteng. E. coli was concentrated from the samples using standard filtration techniques with subsequent incubation on E. coli/coliform chromogenic media to determine the E. coli levels. Bacterial DNA was isolated from bacterial colonies trapped on polyethersulphone membranes after filtration using a celite/guanidium thiocyanate method. A single multiplex PCR (m-PCR) assay was used that targeted the mdh, eaeA, stx1, stx2, st, lt, ial and eagg genes associated with diarrhoeagenic E. coli. The mdh gene was detected in all of the samples even if no culturable E. coli was detected. All the diarrhoeagenic E. coli types were detected in one or more of the raw sewage samples from the various plants. EPEC was present in 20% (2/10) of the samples, EHEC in 50% (5/10), ETEC in 80% (8/10), EIEC in 10% (1/10) and EAEC in 90% (9/10) of the samples. In the case of the primary and secondary treatment only ETEC (5/5; 100%) and EAEC (5/5; 100%) were detected in all of the samples. The results demonstrate that molecular techniques such as PCR have the potential to be used for the monitoring of water samples for the presence of pathogenic E. coli, without the need to culture the organisms.
Keywords: E. coli, multiplex PCR, wastewater treatment plant effluent
Introduction
In South Africa many communities still depend on untreated surface water and ground-water sources for their daily water needs. Water from these sources is often contaminated by faecal pollution from wastewater effluents (Toze, 2004). Wastewater is a matrix consisting of raw sewage and primary, secondary and tertiary treatment effluents (Mara and Horan, 2003). Final effluents are released into receiving waters like dams and rivers. If treatment fails, effluents of poor microbial quality enter public waters (Jagals, 1997).
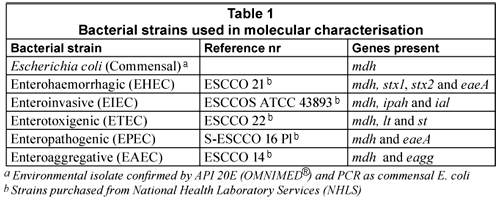
Microbiological indicators have been used for decades to monitor faecal pollution of water (Grabow, 1996; Standard Methods, 2005) as well as the possible presence of other microbiological pathogens (Medema et al., 2003). Escherichia coli is used as an indicator of faecal pollution that originates from human and warm-blooded animals. In the last century, certain E. coli strains were identified as a significant cause of gastro-intestinal disease and recognised as highly versatile pathogens (Nataro and Kaper, 1998). At present, 6 groups of E. coli patho-types have been identified, of which 5 were selected for this study based on their association with diarrhoeal disease in South Africa. These are classified into 5 categories, namely, entero-pathogenic (EPEC), entero-toxigenic (ETEC), entero-invasive (EIEC), entero-aggregative (EAEC) and entero-haemorrhagic (EHEC) (Ashbolt, 2004; Kaper et al., 2004). E. coli strains can thus be either commensal (ComEC; non-diarrhoeagenic) or diarrhoeagenic.
Traditional E. coli detection methods are based on chromogenic and fluorogenic media designed to enumerate ComEC with no relationship to the proportion of diarrhoeagenic E. coli (DEC). All wastewater treatment plants use culture-based methods based on this principle to monitor the water quality. One complication with culture-based methods is their inability to detect E. coli in the viable but non-culturable (VNBC) state (Lleo et al., 2005). VBNC cells are bacterial cells that are metabolically active but cannot be cultured using standard microbiological techniques. These cells are reported to be able to resume active growth when favourable conditions are restored (Chen et al., 2006). It has been reported that pathogenic VBNC cells retain their virulence genes making the water source a potential reservoir of disease (Garcia-Armisen and Servais, 2004).
This inability of traditional culture methods to detect VBNC cells has prompted interest in alternative techniques to monitor the microbiological quality of water (Lleo et al., 2005). Molecular biology techniques, such as the polymerase chain reaction (PCR), have been used to detect DEC types by amplifying specific genes associated with the bacterium's virulence (Kong et al., 2002). To date PCR has mostly been used for the characterisation of isolated E. coli cells or the detection of DEC after sample enrichment. DEC genes targeted with PCR include the eaeA gene for EPEC (Aranda et al., 2004), the lt and st enterotoxin genes for ETEC (Pass et al., 2000), ial gene for EIEC (Paton and Paton, 1998), the eagg gene for EAEC (Kong et al., 2002) and the eaeA, stx1 and stx2 for the detection of EHEC (Moses et al., 2006).
PCR can, however, not be used for the detection of VBNC cells in water samples if the method is used to characterise bacterial isolates obtained with standard culture methods, since the VBNC bacteria cannot be isolated (Chen et al., 2006). The simplest way to overcome this would be to isolate DNA from bacterial cells concentrated from the water samples, which is then used as template for the PCR, circumventing the need for culturability.
The aim of this study was to investigate the use of PCR to detect commensal and pathogenic E. coli concentrated from water samples using membrane filtration. During the study both PCR and culture-based methods were used to detect E. coli in raw sewage, and primary, secondary and tertiary effluents from 6 wastewater treatment plants in Gauteng.
Methodology
Growth and maintenance of reference strains
The E. coli reference strains (Table 1) were cultured on plate count agar (PCA) (Oxoid) and incubated under aerobic conditions at 37ºC for 16 h. If a liquid culture was required, E. coli colonies were inoculated into 5 mℓ nutrient broth and grown overnight at 37°C with mild agitation at 200 r/min.
Sample collection
Samples were collected from 6 wastewater treatment plants in the vicinity of Johannesburg, Gauteng, from August 2006 to December 2007. Thirty-four samples were collected from raw sewage (10), primary treatment effluent (5), secondary treatment effluent (5) and tertiary effluent (14). Water samples were collected in 1 ℓ sampling bottles and kept on ice during transport. Samples were analysed within 3 h of collection.
Microbiological analysis
Microbiological analysis of the samples for E. coli was performed using the standard filtration technique (Standard Methods, 2005). Briefly, 100 mℓ of an appropriate dilution of the water sample was filtered in triplicate onto 0.45-µm gridded nitro-cellulose membrane filters (Merck, Germany). The filters were placed onto selective E. coli/Coliform Chromogenic Media (Oxoid, UK) and the plates were incubated for 24 h at 37ºC. All pink colonies were counted as total coliforms (glucoronidase positive) and all purple colonies were counted as E. coli (β-D-glucoronidase and β-D-galactosidase positive), according to the supplier's instructions. Bacterial counts obtained were expressed as colony forming units in 100 mℓ (cfu/100 mℓ), using the average count of the 3 filters.
Bacterial DNA extraction from wastewater samples
For the DNA extraction, a 100 mℓ sample was filtered onto polyether sulfone (PES) membranes (Microsep (PTY) LTD). DNA was extracted from the trapped bacteria using a modification of the silica/guanidium thiocyanate method reported by Boom et al. (1990) with spin columns prepared as reported by Borodina et al. (2003). The Boom et al. (1990) protocol was followed with 2 exceptions: 250 µℓ 100% (vol/vol) ethanol was added to the lysis buffer, and the celite containing the bound DNA was loaded into the spin columns prior to the washing steps (Borodina et al., 2003). DNA was eluted with 50 µℓ Qiagen elution buffer (Southern Cross Biotechnology®). Controls were included during the DNA extractions and subjected to the same protocol as the samples. The positive control contained 1 mℓ of overnight commensal E. coli liquid culture and the negative control contained 1 mℓ of sterile distilled water.
Multiplex polymerase chain reaction (m-PCR)
The oligonucleotide primers used in this study were synthesised by Inqaba Biotec (Pty.) Ltd. as well as Whitehead Scientific (Pty.) Ltd. The sequences, target genes and expected amplification products are listed in Table 2.
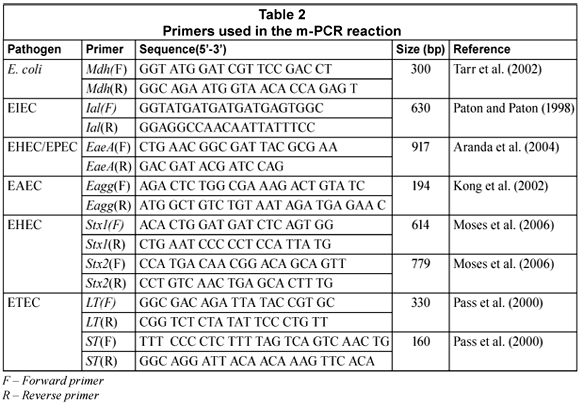
All m-PCR reactions were performed in a Biorad MycyclerTM Thermal cycler in a total volume of 20 mℓ. A m-PCR kit (Qiagen®) was used for the m-PCR protocol. Each reaction consisted of 1X Qiagen® PCR multiplex mix (containing HotstartTaq® DNA polymerase, m-PCR buffer and dNTP mix); 2 µℓ of the primer mixture (0.1 mM of mdh and lt primers Forward (F) and Reverse (R)), 0.2 mM of ial and eagg primers (F and R), 0.3 mM of eaeA and stx2 primers (F and R), 0.5 mM of stx1 and st primers(F and R) (Table 1), 4 mℓ of sample DNA and 4 mℓ PCR grade water. The reactions were subjected to an initial activation step at 95°C for 15 min, followed by 35 cycles consisting of denaturing at 94°C for 45 s, annealing at 55°C for 45 s, extension at 68°C for 2 min and final elongation at 72°C for 5 min (Omar, 2007). Positive and negative controls for the PCR reaction were also included. The positive control contained a mixture of the 5 pathogenic E. coli and commensal E. coli DNA. The negative control contained PCR grade water.
Gel electrophoresis
DNA was analysed on a horizontal agarose slab gel (2.5 % (w/v)) with ethidium bromide (0.5 mg/mℓ) in TAE buffer (40 mM Tris acetate; 2 mM EDTA, pH 8.3). Electrophoresis was performed for 1 to 2 h in electric field strength of 80 V; PCR products were visualised with UV light (Syngene, UK). This procedure was followed for all the experiments except where stated differently. The relative sizes of the DNA fragments were estimated by comparing their electrophoretic mobility with that of the standards run with the samples on each gel, either 1 kB or 100 bp markers (Fermentas, US).
Results and discussion
The microbiological data for the enumeration of E. coli from the samples is shown in Table 3. The average bacterial counts calculated (min; max) for the various samples were 1.94 x 107 cfu/100 mℓ (1.06 x 107 cfu/100 mℓ; 2.34 x 107 cfu/100 mℓ) for the raw sewage (n=10), 9.69 x 106 cfu/100 mℓ (9.43 x 106 cfu/100 mℓ; 9.95 x 106 cfu/100 mℓ) for the primary treatment effluent (n=5), 1.02 x 105 cfu/100 mℓ (8.83 x 103 cfu/100 mℓ; 1.96 x 105 cfu/100 mℓ) for the secondary treatment effluent (n=5) and 1.59 x 104 cfu/100 mℓ (0 cfu/100 mℓ; 7.15 x 104 cfu/100 mℓ) for the tertiary effluent (n=14). The breakdown of these counts per treatment plant is also shown in Table 3.
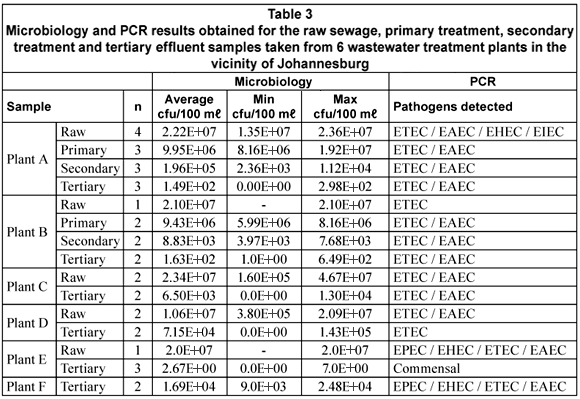
As expected the number of E. coli present in the samples declined from raw sewage (starting material) up to the tertiary effluent (final product). Bacterial counts for the tertiary effluents did however cover a broad range from 2.67 x 100 cfu/100mℓ up to 7.15 x 104 cfu/100mℓ, which provided a range of contexts in which to test PCR as an alternative for the detection and characterisation of E. coli strains present.
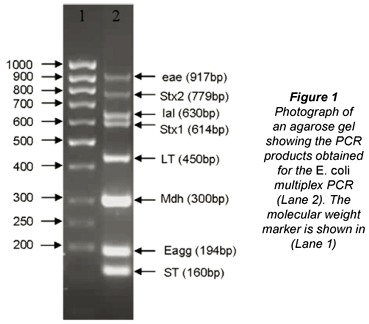
The m-PCR used for the sample analyses targeted 7 of the virulence genes mostly associated with EHEC (stx1, stx2, eaeA), EPEC (eaeA), ETEC (lt, st), EIEC (ial), EAEC (eagg) as well as the mdh gene found in both commensal and diarrhoeagenic E. coli (Omar, 2007). An example of the complete m-PCR is shown in Fig. 1.
Individual E. coli colonies isolated on selective media were not tested using the m-PCR. This was avoided as pathogenic strains would represent only a small proportion of the total E. coli community.
The initial PCR results obtained showed PCR inhibition in 21% (7/34) as indicated by the absence of an mdh product. Since all the samples had culturable E. coli it was expected that at least the mdh gene would be detected in the samples. The mdh gene could be detected in all 7 samples after the DNA was diluted 5 times, lifting the effect of the PCR inhibitors. The dilution of DNA should always only be considered as a last resort since this can possibly limit the detection of DEC in the sample, especially if the DEC is present in very low numbers. Due to the nature of the samples it was, however, necessary.
The general distribution of the DEC types in the various stages of the wastewater treatment plants is given in Table 4. All the DEC types were detected in 1 or more of the raw sewage samples from the various plants. EPEC was present in 20% (2/10) of the samples, EHEC in 50% (5/10), ETEC in 80% (8/10), EIEC in 10% (1/10) and EAEC in 90% (9/10) of the samples (Table 4). In the case of the primary and secondary treatments only ETEC (5/5; 100%) and EAEC (5/5; 100%) were detected in all of the samples. All the DEC types except for EIEC could be detected in the tertiary effluent samples. The presence of EHEC (2/14; 14.3%) and EPEC (2/14; 14.3%) was however restricted to Plant F (Table 5). The tertiary effluent of Plant F not only showed the presence of high E. coli counts but also the presence of EHEC, EPEC, EAEC and ETEC in both samples tested.
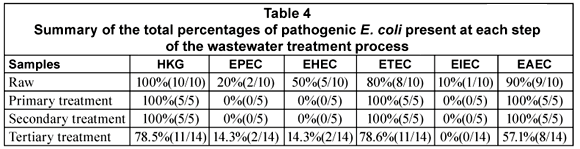
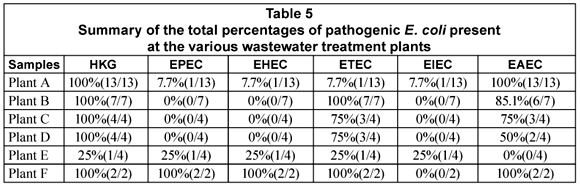
In the case of Plant A, ETEC and EAEC could be detected in the raw sewage, primary and secondary treatment effluents as well as the tertiary effluent. The pathogens were, however, not present in all of the samples taken from each point. The same trend was seen with Plant B with the exception that ETEC was detected in all of the samples tested.
Although it was expected that the mdh gene would at least be detected in the raw sewage and primary and secondary treatment effluents due to the high number of E. coli present, the ability of the PCR method in detecting not only mdh but also virulence genes in the tertiary effluents was very encouraging. The mdh gene could even be detected in samples with low or no E. coli detected using culture-based methods. It should, however, be noted that although DEC were detected with the PCR, especially in the tertiary effluent, no conclusions can be made about the viability of the bacterial cells detected. This is mainly due to the fact that PCR will amplify genes from the DNA of both viable and dead cells.
Conclusion
PCR has the potential to be used for the monitoring of water samples for the presence of pathogenic E. coli without the need to culture the organisms. This method can be extended to test for other bacterial pathogens such as Vibrio cholerae in water samples if an appropriate PCR protocol is available. For this process to be more efficient more research is needed into the DNA extraction method to ensure that no PCR inhibitors are present after DNA extraction, especially when working with samples such as raw sewage. This will ensure that no DNA dilutions are required that might reduce the chance of detecting bacterial pathogens in the samples.
It is important to consider that although the pathogens were detected with the m-PCR, the genes could have been amplified from DNA recovered from dead and/or viable cells. Since humans might use the water downstream of the treatment plant, there is a need to better understand what pathogens are released and whether they are still virulent. Olivier et al. (2005) showed that E. coli and Salmonella typhimurium can enter the viable but non-culturable state if secondary treated wastewater is chlorinated, leading to a drastic underestimation of the actual quality of the water released into the environment.
Acknowledgements
This project was completed with funding obtained from the University of Johannesburg and the National Research Foundation. We would like to express our sincere gratitude to the managers at the wastewater treatment plants, for allowing us to collect the samples.
References
ARANDA KRS, FAGUNDES-NETO U and SCALETSKY (2004) Evaluation of multiplex PCR's for diagnosis of infection with diarroeagenic Escherichia coli and Shigella spp. J. Clin. Microbiol. 42 5849-5853. [ Links ]
ASHBOLT JH (2004) Microbial contamination of drinking water and disease outcomes in developing regions. Toxicol. 198 229-238. [ Links ]
BOOM R, SOL CJA, SALIMAN MMM, JANSEN CL, WERTHEIM-VAN DILLEN PME and VAN DER NOORDAA J (1990) Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28 495-503. [ Links ]
BORODINA TA, LEHRACH H and SOLDATOV AV (2003) DNA purification on homemade silica spin-columns. Anal. Biochem. 321 135-137. [ Links ]
CHEN YC, HIGGINS MJ, MAAS NA and MURTHY SN (2006) DNA extraction and Escherichia coli quantification of anaerobically digested solids using the competitive touchdown PCR method. Water Res. 40 3037-3044. [ Links ]
GARCIA-ARMISEN T and SERVAIS P (2004) Enumeration of viable E. coli in rivers and wastewaters by fluorescent in situ hybridization. J. Microbiol. Methods 58 269-279. [ Links ]
GRABOW WOK (1996) Waterborne diseases: Update on water quality assessment and control. Water SA 22 (2) 193-202. [ Links ]
JAGALS P (1997) Stormwater runoff from typical developed and developing South African urban developments: Definitely not for swimming. Water Sci. Technol. 35 (11-12) 133-140. [ Links ]
KAPER JB, NATARO JP and MOBLEY HLT (2004) Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 123-140. [ Links ]
KONG RYC, LEE SKY, LAW TWF, LAW SHW and WU RSS (2002) Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 36 2802-2812. [ Links ]
LLEO MM, BONATO B, TAFI MC, SIGNORETTO C, PRUZZO C and CANEPARI P (2005) Molecular vs culture methods for the detection of bacterial faecal indicators in groundwater for human use. Appl. Microbiol. 40 289-294. [ Links ]
MEDEMA GJ, PAYMENT P, DUFOUR A, ROBERTSON W, WAITE M, HUNTER P, KIRBY R and ANDERSON Y (2003) Safe drinking water: An ongoing challenge. In: Dufour A, Snozzi M, Koster W, Bartram J, Ronchi E and Fewtrell L (eds.) Assessing Microbial Safety of Drinking Water: Improving Approaches and Methods. Organisation for Economic Co-operation and development (OECD), World Health Organisation (WHO). IWA, London. [ Links ]
MARA D and HORAN N (2003) The Handbook of Water and Wastewater Microbiology. Academic Press, USA. 193 pp. [ Links ]
MOSES AE, GARBATI MA, EGWU AO and AMEH EJ (2006) Detection of E. coli 0157 and 026 serogroups in human immunodeficiency virus-infected patients with clinical manifestation of diarrhoea in Maiduguri, Nigeria. Res. J. Med. Med. Sci. 1 (4) 140-145. [ Links ]
NATARO JP and KAPER JB (1998) Diarrhoeagenic Escherichia coli. Clin. Microbiol. Rev. 11 (1) 142-201. [ Links ]
OLIVIER JD, DAGHER M and LINDEN K (2005) Induction of Escherichia coli and Salmonella typhimirium into the viable but non-culturable state following chlorination of wastewater. J. Water Health 3 (3) 249-256. [ Links ]
OMAR KB (2007) Determining the pathogenicity and quantities of Escherichia coli in selected South African water types using molecular biology techniques. M.Tech Biotechnology Thesis, University of Johannesburg. [ Links ]
PASS MA, ODEDRA R and BATT RM (2000) Multiplex PCR for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38 2001-2004. [ Links ]
PATON AW and PATON JC (1998) Detection and characterization of shiga toxigenic Escherichia coli by using multiple PCR assay for stx1, stx2, eaeA, EHEC hlyA, rfb. J. Clin. Microbiol. 36 598-602. [ Links ]
STANDARD METHODS (2005) Standard Methods for the Examination of Water and Wastewater (21st edn.). Eaton AD, Clesceri LS, Rice EW and Greenberg AE (eds.). American Public Health Association, Washington DC, USA. [ Links ]
TARR CL, LARGE TM, MOELLER CL, LACHER DW, TARR PI, ACHESON DW and WHITTMAN TS (2002) Molecular characterization of a serotype 0121:H19 clone, a distinct shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70 (12) 6853-6859. [ Links ]
TOZE S (2004) Reuse of effluent water – benefits and risks, new directions for a diverse planet. Proc. 4th International Crop Science Congress, 26 September 2004, Brisbane, Australia. [ Links ]
This paper was originally presented at the 2010 Southern African Young Water Professionals Conference, Pretoria, 19-20 January 2010
* To whom all correspondence should be addressed
+2711 559 6324; fax: +2711 559 6329
e-mail: kousaro@uj.ac.za














