Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Water SA
On-line version ISSN 1816-7950
Print version ISSN 0378-4738
Water SA vol.36 n.2 Pretoria Jan. 2010
Optimisation of methods for the collection and detection of bacterial pathogens from diarrhoeal human faecal samples using a novel stool collection kit
SIK MietaI; N PotgieterII; MD SobseyIII; TG BarnardI, *
IWater and Health Research Unit, University of Johannesburg, PO Box 17011, Doornfontein 2028, South Africa
IIDepartment of Microbiology, University of Venda, Private Bag X5050, Thohoyandou, 0950, Limpopo Province, South Africa
IIIUniversity of North Carolina, CB# 7431, Rosenau Hall, Room 148,Chapel Hill, NC 27599-7431, USA
ABSTRACT
Bacterial pathogens such as Escherichia coli, Salmonella-, Shigella- and Vibrio species are known to be common causative agents for diarrhoeal disease in humans. This study aimed to develop a culture-independent PCR assay for the detection of bacterial pathogens present in human faecal samples collected using a less intrusive faecal collection technique, the Bio-wipe kit. A multiplex-PCR (m-PCR) was optimised targeting the E. coli mdh gene, the Salmonella IpaB gene, the Vibrio sodB gene, and the Ial and IpaH genes present in entero-invasive E. coli and Shigella spp. The influence of the DNA extraction method, and sensitivity and specificity of the m-PCR and the Bio-wipe storage conditions on the detection of the bacterial pathogens was investigated. A guanidium thiocyanate DNA extraction method used with laboratory-prepared spin columns could successfully extract DNA from 93% of the samples analysed. The m-PCR could successfully identify and differentiate between the various pathogens tested and was specific for the selected pathogens. Faecal matter was successfully recovered from used Bio-wipes and the bacterial DNA could be detected from these samples at concentrations of 10 cfu. Bacterial DNA could be recovered from the Bio-wipes 5 to 10 d after use when the Bio-wipes were stored at 30°C and 14 d after usage when stored at ambient temperature. Thus the Bio-wipe kit, along with the m-PCR, can be used for collection and detection of bacterial pathogens during outbreaks and in rural settings.
Keywords: Bio-wipe kit, bacterial pathogens, faecal matter, PCR, DNA extraction
Introduction
The human intestinal tract is home to a complex community of microbial species which serve as markers of gastro-intestinal health (Zhang et al., 2006). The occurrence and distribution of bacterial pathogens causing diarrhoea in humans has been shown in various studies (Reyes et al., 2009). Bando et al. (2009) reported that a high incidence of infantile diarrhoea is associated with atypical enteropathogenic E. coli (EPEC); in addition, data published by Khan et al. (2008) indicated that Vibrio cholerae, Shigella and Salmonella spp. are commonly associated with neonatal diarrhoea in Bangladesh.
The approach commonly used for the identification and detection of these pathogens is based on either bacterial isolate characterisation or the presence of pathogens in a mixed sample (Dedeić-Ljubović et al., 2009; Estrada-Garcia et al., 2009; Silva et al., 2008). The first problem encountered with isolate characterisation, as reported by many researchers, is the under-detection of the causative agent due to the typically small number of bacterial colonies tested per sample (Galbadage et al., 2009; Jafari et al., 2009; Ajjampur et al., 2008). A second problem is the inaccessibility of laboratories to rural clinics, preventing the efficient isolation and detection of bacterial pathogens.
The detection of pathogens within mixed samples has become the preferred identification route and many studies employ molecular techniques such as the polymerase chain reaction (PCR), which identifies the causative agents of diarrhoeal disease (Ram et al., 2009; Usein et al., 2009; Ajjampur et al., 2008; Meraz et al., 2008). PCR techniques are culture-independent therefore the samples collected can be stored for longer periods prior to analysis while the bacterial pathogens can still be detected.
The problem encountered, especially in rural areas, is the collection and storage of samples. Faecal samples from infected individuals are typically collected in sterile universal containers or with rectal swabs, especially in rural areas where no clinics or hospitals are available. The collection of faecal samples can negatively influence the diagnosis and monitoring of diarrhoea due to factors such as delays in obtaining samples from individuals and refusal of individuals, due to their ethnic and social beliefs, to provide faecal matter (Duncker et al., 2007).
To improve the faecal collection technique, a new method, termed the Bio-wipe kit, was obtained from the American Centre for Disease Control through Prof. Mark Sobsey. The Bio-wipe is used in the same manner as toilet paper making it a less intrusive collection technique. The aim of this study was to test the Bio-wipe kit, combined with PCR, for the collection of faecal samples from individuals and detection of diarrhoeal pathogens. This was achieved by evaluating 3 DNA isolation methods to determine their relative efficiency and DNA yield, developing and optimising a single multiplex PCR assay to detect housekeeping and/or virulence genes for E. coli, Salmonella, Shigella and V. cholerae species, and, lastly, determining the influence of storage conditions on bacterial DNA purified from faecal matter collected onto the Bio-wipe.
Materials and methods
Bacterial strains used
Bacterial strains used for the experimental work (Table 1) were obtained from the National Health Laboratory Services (NHLS), American Type Culture Collection (ATCC) and the National Collection of Type Cultures (NCTC). All bacterial strains were stored at -70°C in MicrobankTM cryo-vials (Pro-Lab Diagnostics, USA). The strains were grown on Plate Count Agar (PCA; pH 7.0) (Merck, Germany) or Nutrient Agar (pH 7.4) (Oxoid, England) under aerobic conditions at 37°C for 16 to 24 hours. Prior to spiking of faecal specimens, Escherichia coli, Vibrio cholerae non-01, Salmonella typhimurium and Shigella dysenteriae were inoculated into nutrient broth (pH 7.4) (Oxoid, England) and incubated at the above-mentioned conditions with agitation at 200 r/min.
Human faecal samples
Faecal specimens were obtained from the Microbiology Department of the National Health Laboratory Services (NHLS). Ethical approval was given by the University of Johannesburg Academic Ethics Committee and the required patient confidentiality was maintained as the identities of the individuals were unknown. Thirty specimens were randomly selected to represent various stool compositions. The faecal matter was received in sterile collection containers and was stored at 4°C prior to weighing. Approximately 0.2 g of each of the faecal samples were suspended in 1.5 mℓ phosphate buffered saline (pH 7.4) (Sigma Aldrich, USA) containing 0.1% (vol/vol) Tween buffer (Merck, Germany) and homogenised by vortexing. This suspension was aliquoted into clean 2 mℓ eppendorf tubes and stored at -20°C until it was needed.
The Bio-wipe kit
Materials and assembly of the Bio-wipe kit
The Bio-wipe kit consists of a 10 x 10 cm square absorbent material with an orange plastic backing (Fisher Scientific, USA). It was placed in a sterile re-sealable bag. Polyester batting material was cut to the size of the Bio-wipe and placed in a 2nd re-sealable sterile bag. The batting material was soaked with 7 mℓ storage media (sodium thioglycolate, Na2HPO4.7H2O, NaCl, 1% CaCl2 and agar). Both the Bio-wipe and the batting material were placed into an envelope with a unique identification number (for control purposes) prior to use.
Use of Bio-wipes in the laboratory
All Bio-wipes were freshly prepared the day before use. The faecal matter was smeared evenly onto un-used Bio-wipes, placed onto the batting material containing the storage media, folded together and placed into the sterile re-sealable bag. The Bio-wipe was kept at the conditions required for the specific experiments.
Recovery of faecal material from Bio-wipes
Faecal material was recovered from the Bio-wipes in a Bio-safety Cabinet Level 2 (ESCO Biotech, USA). The surfaces was disinfected before and after recovering faecal matter from each Bio-wipe and 2 pairs of clean gloves were worn before handling each Bio-wipe. The Bio-wipe containing the faecal matter and the batting material was removed from the envelope and was placed into a 150 mm diameter sterile glass Petri dish (LASEC; South Africa) with the faecal matter side facing upwards. Approximately 6 to 10 mℓ of PBS + 0.1% Tween buffer (vol/vol) was pipetted onto the Bio-wipe. The faecal matter was suspended from the Bio-wipe by repeatedly pipetting ~3 mℓ of the buffer from the Petri dish onto the Bio-wipe, until most of the faecal matter was dislodged from the Bio-wipe surface. The buffer containing the faecal matter was aliquoted into 2 mℓ cryo-vials and stored at -20°C.
Comparison of DNA extraction methods
The guanidinium thiocyanate (GuSCN) DNA extraction method (Boom et al., 1990), guanidinium thiocyanate DNA extraction method with alpha-casein (Boom et al.,1999) and the commercial QIAamp® DNA stool mini kit were selected for the extraction of bacterial DNA from faecal matter.
Guanidinium thiocyanate DNA extraction method without alpha casein
An adaptation of the protocol reported by Boom et al. (1990) was used for this study. Changes included the use of the stool suspension as starting material that was vortexed, homogenised and centrifuged at 13 000 r/min for 5 min after the samples were thawed. Furthermore the pellet was used for DNA extraction and the supernatant discarded. The pellet was suspended in 700 µℓ L6 lysis buffer (5.25M guanidium thiocyanate, 0.1 M tris hydrochloride, 0.2 M EDTA and 1.3% (weight/vol) Triton X-100) and incubated at 70°C for 10 min. A volume of 250 µℓ 100% (vol/vol) ethanol was added to this mixture and further incubated at 56°C for 10 min. The celite solution (50 µℓ) was added and incubated at room temperature for 10 min (with occasional mixing of the mixture). A sterile spin column, prepared according to the method published by Borodina et al. (2003), was placed into a sterile 2 mℓ microfuge tube and the mixture loaded into the column. The mixture was loaded by adding approximately 500 µℓ of the lysis mixture into the column followed by centrifugation at 13 000 r/min for 30 s to separate the buffer from the celite. This step was repeated twice until all of the lysis mixture was loaded into the column. The column was washed twice with 400 µℓ wash buffer (5.25M guanidium thiocyanate and 0.1 M tris hydrochloride) and twice with 400 µℓ of a 70% (vol/vol) ethanol solution, separating the liquid and solid phase each time by centrifugation at 13 000 r/min for 30 s. The last wash step was followed by a 2 min centrifugation step at 13 000 r/min to ensure that all the ethanol was removed from the column. Columns were transferred into clean sterile 1.5 mℓ microfuge tubes and 100 µℓ elution buffer (AE buffer, Qiagen) was added to the columns and incubated for 2 min at 56°C. DNA was eluted from the columns by centrifugation for 2 min at 13 000 r/min after which the columns were discarded. DNA containing AE buffer was collected into the 1.5 mℓ microfuge tube. A negative control was included by performing the DNA extraction methods with only the DNA extraction reagents. A positive control was prepared by extracting DNA from 1.5 mℓ culture of the bacteria.
Guanidinium thiocyanate DNA extraction method with alpha casein
In order to prepare buffer L7A (Boom et al., 1999), 1 mg/mℓ ά-casein (Sigma-Aldrich, Germany) was added to buffer L6 (lysis buffer); 200 µℓ of the faecal suspension (as described above) was then added to a microfuge tube containing 1000 µℓ L7A buffer and 60 µℓ celite, vortexed and incubated at room temperature for 10 min. The remainder of the protocol was followed as described above.
QIAamp® DNA stool mini kit method
The protocol was followed as stipulated by the supplier using 180 to 220 mg of each faecal specimen (Qiagen Handbook, 2001).
Multiplex PCR primers and method
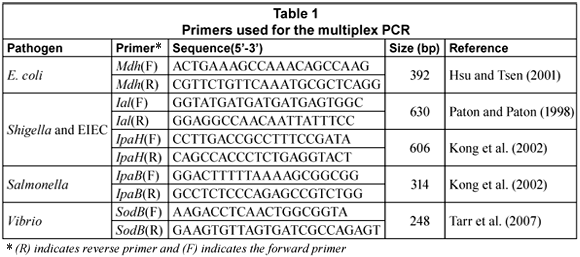
The multiplex PCR (m-PCR) targeted the mdh gene for E. coli (Hsu and Tsen, 2001), the IpaB gene for Salmonella (Kong et al., 2002), the IpaH and Ial genes for Shigella and Entero-invasive E. coli (Kong et al., 2002; Paton and Paton, 1998) and the sodB gene for Vibrio spp (Tarr et al., 2007).
Primers (Table 1) were obtained from Whitehead Scientific. PCR reactions were performed in a Biorad MycyclerTM Thermal cycler in a 20 µℓ reaction mixture that consisted of 1 X QIAGEN multiplex PCR master mix (Qiagen, Germany). To this, 100 pmol of each primer, 2 µℓ of the DNA template and PCR grade water were added to make up the final volume of 20 µℓ. To create a positive control, 2 µℓ of a V. cholerae O1, entero-invasive E. coli (EIEC), S. dysenteriae and S. typhimurium DNA mixture was included; to create a negative control template DNA was excluded.
The amplification cycle conditions consisted of an initial activation at 95°C for 15 min, followed by 35 cycles of denaturation at 94°C for 45 s; annealing at 57°C for 45 s and extension at 72°C for 1 min. This was followed by a final extension step at 72°C for 5 min.
Electrophoresis and visualisation of DNA
DNA was analysed in a horizontal agarose slab gel (2.5% (weight/vol)) containing ethidium bromide (0.5 µg/mℓ) in TAE buffer (40 mM tris-acetate; 2 mM EDTA, pH8.3). The agarose gel was electrophoresed for 1 to 2 h at 80 to100 V. The DNA was visualised with UV light (Gene Genius Bio Imaging System, Vacutec®). The relative sizes of the DNA fragments were estimated by comparing their electrophoretic mobility with 100 bp markers (Fermentas O' GeneRuler DNA ladder; Canada).
Detection limits and specificity of the assay
The specificity of the multiplex PCR was confirmed against other related and unrelated enteric bacterial DNA. The sensitivity of the m-PCR was determined by measuring the optical densities (OD550nm) of overnight bacterial suspensions to standardise the bacterial concentrations. The cell suspensions were serially diluted 10-fold to obtain a cell suspension range of 1 x 100 to 1 x 106 cfu/mℓ (Hong, 2007). The DNA was extracted from 2 mℓ of the diluted bacterial suspensions and the m-PCR performed.
Optimising storage conditions for the Bio-wipe kit
To investigate the optimal storage conditions for the Bio-wipe samples, bacterial suspensions were grown as described previously and were diluted 10-fold. The faecal samples were mixed with 1 mℓ of 6.4 x 1010 cell suspensions and the faecal matter was smeared onto the Bio-wipes. The desired amount of Bio-wipes was stored at either room temperature, 4°C or 30°C for periods of 3 h, 1 d, 3 d, 7 d, 14 d, 21 d and 30 d. After the incubation period, the faecal matter was recovered from each Bio-wipe; the DNA was extracted using the GuSCN without α-casein DNA extraction method and quantified using the QubitTM Quantification Platform Fluorometer (Invitrogen; USA), and the multiplex PCR was performed. The experiments were performed in triplicate.
Results and discussion
Multiplex PCR
The m-PCR was designed to target genes specific to the 4 entero-pathogenic bacteria selected for this study and can be used as a screening procedure for all faecal samples. The mdh gene encodes for malic acid dehydrogenase, a housekeeping enzyme of the citric acid cycle, which is reportedly found in all E. coli strains (Hsu and Tsen, 2001). Identifying the mdh gene confirms the presence/absence of both commensal and pathogenic E. coli in the faecal matter. The Ial and IpaH virulence genes, as stated previously, are present in both EIEC and Shigella spp. (Paton and Paton, 1998; Kong et al., 2002). The Ial gene is not only located on the chromosome of EIEC, but is also located on the inv plasmid of the Shigella spp. (Hsu and Tsen, 2001). It has been reported that the detection of the Ial gene may give false negative results as the inv plasmid is prone to loss/deletions, unlike the IpaH gene which is present on both the chromosome and the inv plasmid of Shigella spp., making it a more stable gene for the detection of Shigella spp. (Hsu and Tsen, 2001).
For the detection of Salmonella spp. the IpaB gene, which is a virulence gene found on the invasion plasmid of Salmonella spp, was selected for the PCR as it is reportedly present in most Salmonella strains (Kong et al., 2002). The sodB gene is a conserved housekeeping gene present in all Vibrio spp.; thus targeting the housekeeping gene confirms the presence of all Vibrio spp. (Tarr et al., 2007).
Although various PCR methods have been reported for the simultaneous detection of various combinations of diarrhoeagenic bacteria (Stone et al., 1994; Houng et al., 1997; Collins et al., 2001; Gentry-Weeks et al., 2002; Kong et al., 2002; Gubala, 2006), the m-PCR used in this study was designed for the simultaneous detection of E. coli, Vibrio, Salmonella and Shigella species, which are the most commonly encountered bacterial pathogens in South Africa.
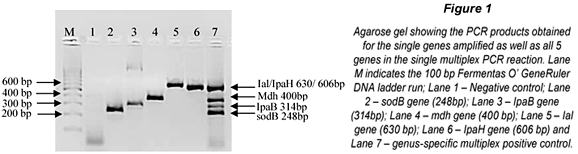
The m-PCR assay was successfully optimised as shown in Fig. 1. In the combined multiplex reaction only 4 bands are visible. This is due to the similar molecular weights of the Ial (630bp) and IpaH (606bp) genes which can be seen in Lanes 5 and 6 of Fig. 1. Further effort to separate the bands was not required as the Ial and IpaH genes encode for the invasion and colonisation of intestinal cells in both EIEC and Shigella spp. and the presence of either one indicates the presence of EIEC and/or Shigella spp (Paton and Paton, 1998; Kong et al., 2002).
Detection limits and specificity
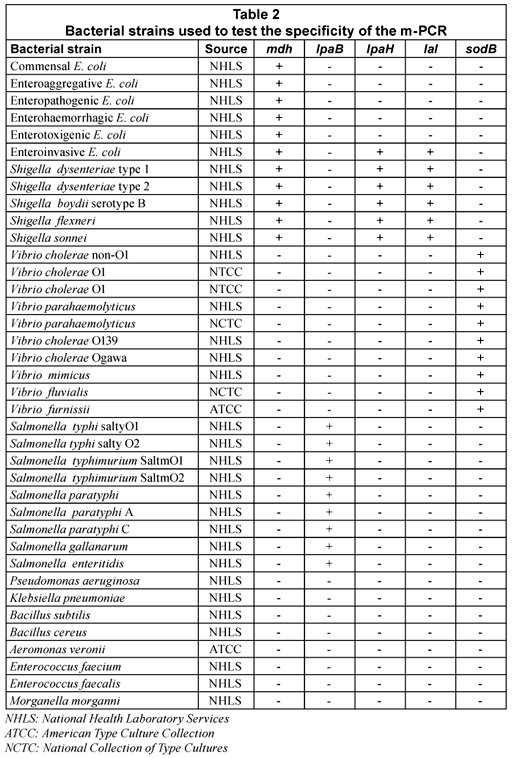
The specificity of the m-PCR was assessed by testing the assay on 38 bacterial strains, which included commensal and pathogenic E. coli strains, Shigella spp., Salmonella spp., Vibrio spp. and other strains of the Enterobacteriaceae family such as Klebsiella, Aeromonas, etc. (Table 1). Results showed that all the E. coli, Salmonella, Shigella and Vibrio spp. strains generated positive results for the specific genes, whereas DNA from the other enterobacteriaceae strains that were tested were not amplified by the m-PCR. The sodB gene was detected in all Vibrio strains tested but was also detected in Aeromonas veronii in 1 test. This gene could, however, not be detected in further repeated analysis of Aeromonas veronii. The false positive result detected for the sodB gene for the A.veronii isolate could be due to the similarities between Vibrio spp. and Aeromonas spp (Abbott et al., 1998). It can thus be concluded that the m-PCR is specific for detecting E. coli, Salmonella, Shigella and Vibrio spp. from faecal samples.
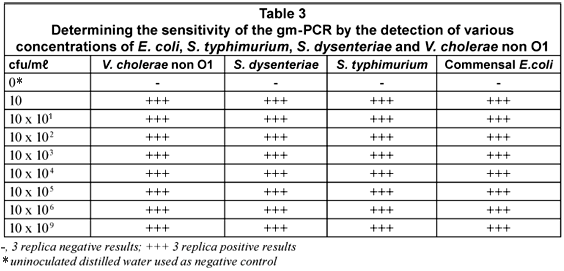
The sensitivity of the m-PCR was measured against bacterial cultures of E. coli, S. typhimurium, S. dysenteriae and V. cholerae non O1 ranging from 1 x 100 to 1 x 109 cfu/mℓ in sterile distilled water. The detection limit for each of the selected pathogens was 10 cfu for the m-PCR assay (Table 3).
Comparison of DNA extraction methods
The purpose of the study was to test 3 DNA extraction methods, two of which were reported by Boom et al. (1990) and Boom et al. (1999), respectively, and a commercially available QIAamp® DNA stool mini-kit that has been optimised for the isolation of bacterial DNA from human faecal matter. DNA could be extracted from 67% of the unspiked samples with the GuSCN (non ά-casein) method compared to 27% of the unspiked faecal samples using the GuSCN including ά-casein method. The QIAamp® DNA stool mini-kit was only successful in isolating DNA from 6% of the unspiked samples. Upon spiking of the faecal samples the DNA recovered with the GuSCN (non ά-casein) method decreased in comparison to the DNA recovered using the GuSCN including ά-casein method. However the overall DNA yields for the GuSCN (non ά-casein) DNA extraction method remained higher than for the other 2 methods (Table 4). The PCR results obtained from the unspiked samples showed that DNA could be detected from 93% (28/30) of the samples tested using the GuSCN (non ά-casein) extraction method, 93% (28/30) of samples using the GuSCN with ά-casein extraction method and 46% (14/30) of samples using the QIAamp® DNA stool mini kit (Table 4). These results showed that some of the PCR positive samples had undetectable DNA concentrations.
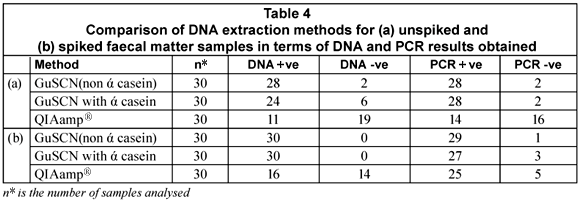
The findings indicated that the DNA yields for the GuSCN (non ά-casein) method (Boom et al., 1990) were better when compared to the other 2 methods. The function of ά-casein in the GuSCN with ά-casein method was to prevent the binding of PCR inhibitors to the silica particles during DNA extraction. Results obtained from the study indicated that the presence of ά-casein did not impact greatly on the recovery rate of bacterial DNA from the faecal matter samples. The significant variability in the total DNA yields for the extraction methods may be as a result of variations in the composition of faecal matter, the fact that the initial amount of starting material differs for each method, and the DNA quantification kit used not having a low enough detection limit.
The QIAamp® DNA stool mini-kit has been reported to successfully extract bacterial DNA from faeces, in comparison to other commercial kits available (McOrist et al., 2002; Folton et al., 2005; Nechvatal et al., 2008). However, in this study it was noted that the QIAamp® DNA stool mini-kit did not perform as well in terms of the DNA yield when compared to the method published by Boom et al. (1990). This may be due to the fact that the commercial kit was designed and optimised for general laboratory use and not for diagnostic use with human stool samples. The DNA binding system employed by the QIAamp® DNA stool mini-kit is limited to adsorption onto a QIAamp® silica membrane, whereas the GuSCN-based methods use silica particles, in the presence of the chaotrophic salt guanidinium thiocyanate (Boom et al., 1990), as well as the filter membrane in the spin column to capture DNA.
Optimising storage conditions for the Bio-wipe kit
Before the Bio-wipes can be implemented in rural areas the influence of the storage conditions on the detection of the bacterial pathogens has to be understood. The environmental temperatures in the rural areas where the Bio-wipes will be implemented range from 20°C in the winter months to 30 to 37°C in the summer months. The rural areas are not visited daily by nurses who collect and send the samples to the appropriate laboratories. The optimal storage conditions were determined in terms of the DNA yield and the presence of the genus/species-specific virulence genes was determined by visualising the amplified PCR products.
The results presented (Fig. 2) show that bacterial DNA was easily recovered from Bio-wipes stored at ambient temperature. This was expected as the bacteria were kept in favourable conditions (optimal temperature) and the storage media provided the bacteria present with the required nutrients. However, when the Bio-wipes were stored for longer than 2 weeks prior to analysis, the data showed a decline in the recovery of bacterial DNA. Ambient temperature is the most convenient temperature for storage therefore the Bio-wipes can be kept in any South African environment where the temperature ranged between 20 and 25°C.
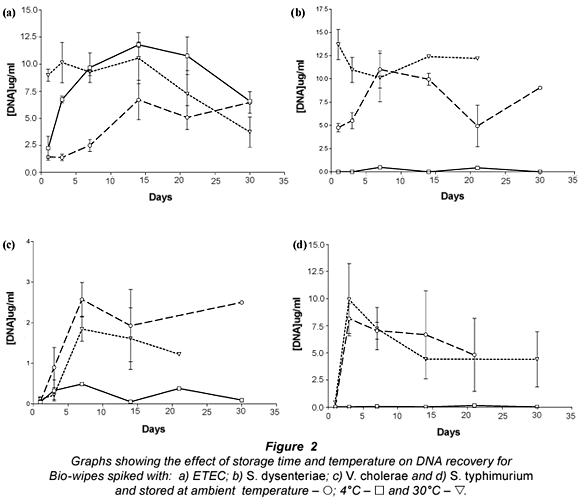
Storage of the Bio-wipes at 4°C showed that lower levels of bacterial DNA were recovered than for Bio-wipes stored at ambient temperature and 30°C. This was expected as lower temperatures slow down the metabolic processes of bacteria leading to a decrease in the rate of cell division (Brözel and Cloete, 1991). The E. coli DNA yield from the Bio-wipes peaked in the samples stored for 14 d; thereafter there was a steady decline. Analysis of the DNA recovery for S. dysenteriae, S. typhimurium and V. cholerae non O1 (Fig. 2b-d) showed that the DNA yield did not exceed 17 ug/mℓ from the Bio-wipes that were spiked with pure cultures.
The results presented for Bio-wipes stored at 30°C indicated large variability in the recovery rate of bacterial DNA. The large variation may be due to the rapid growth of bacteria when the Bio-wipes were stored at higher temperatures, as most of the bacteria present in faecal matter are thermophilic. There was a high DNA yield when the Bio-wipes were analysed after 5 to 10 d of incubation. There was a major decrease in the DNA yield after 14 d of incubation, which may have resulted from the nutrients being depleted and thus a decline in bacterial cell survival. The optimal storage time for the Bio-wipes to obtain accurate data was 14 d, after which the faecal matter could not be recovered from the majority of the Bio-wipes due to fungal contamination.
The selected genus-specific target genes were detected from all the Bio-wipes that were analysed on Day 7 of incubation at ambient temperature. From Fig. 2b-d it is noted that there was more variability in the recovery of DNA from the Bio-wipes spiked with S. dysenteriae, S. typhimurium and V. cholerae non O1 species. The PCR results for the Bio-wipes stored at 4°C showed that the virulence genes for ETEC could not be detected in any of the Bio-wipes; thus ETEC identification was not possible from the Bio-wipes stored at 4°C. When analysing the Bio-wipes that were stored at 4°C the bacterial cells may have shed the plasmid-borne virulence genes and thus were undetectable. The bacterial cells discard plasmids which contain virulence genes that are not needed by the cells during unfavourable conditions, in an attempt to conserve energy resources that are used for survival (Jagals et al., 2006). Alternatively this result may have been due to an experimental flaw at the time of the study.
The virulence genes for S. dysenteriae and S. typhimurium were detected from all the Bio-wipes. However the genus- specific gene for V. cholerae non O1 was not detected from any of the Bio-wipes stored at 4°C. This may have been due to competition with other bacteria present at higher concentrations. For the Bio-wipes incubated at 30°C all the virulence genes were detected.
Conclusion
It can be concluded from the results presented that the Bio-wipe kit and PCR will be a valuable tool provided that certain critical factors and shortcomings are kept in mind. The most notable of these is that the bacterial pathogens may not be isolated with culture-based methods as they may be in the viable but non-culturable state. This is mainly because the system was developed for environments where samples cannot be transported immediately to a laboratory. The Bio-wipe should in general not be stored for more than 2 weeks before it is transported to the laboratory, but it can be kept at elevated temperatures and still result in the successful detection of the bacterial pathogens.
It is evident that there is variability in the total DNA yield for the extraction methods, which is partly due to intra-specimen variability, as faeces are heterogeneous biological materials (McOrist et al., 2002). It can be concluded that the GuSCN (non ά-casein) method is a suitable and cost-effective alternative to the QIAamp® mini-stool kit for the isolation of bacterial DNA from human faecal matter. The m-PCR assay is a sensitive and reliable method to identify E. coli, Salmonella, Shigella and Vibrio spp. from faecal samples and can assist in the rapid identification of the causative agents of diarrhoea. Since the proposed PCR methods are tested against the total population of bacteria present, instead of testing 5 to 10 bacterial colonies per sample, the probability of missing the causative agent is reduced. One drawback of the PCR method is that no statement can be made about the viability of the pathogens detected, but current research is aiming to address this problem.
The use of a combination of the Bio-wipe kit and PCR, for the collection and detection of the bacterial pathogens in stool samples, promises to be a valuable tool in the investigation of diarrhoeal outbreaks or in household intervention studies where diarrhoea is used as an indicator of the effectiveness of an intervention.
Acknowledgements
The authors wish to thank the National Research Foundation of South Africa for funding this project as well as the NHLS for providing the faecal samples for the study.
References
ABBOTT SL, SELI LS, CATINO M (jnr), HARTLEY MA and JANDA JM (1998) Misidentification of unusual Aeromonas species as members of the genus Vibrio: a continuing problem. J. Clin. Microbiol. 36 1103-1104. [ Links ]
AJJAMPUR SS, RAJENDRAN P, RAMANI S, BANERJEE I, MONICA B, SANKARAN P,ROSARIO V, ARUMUGAM R, SARKAR R, WARD H and KANG G (2008) Closing the diarrhoea diagnostic gap in Indian children by the application of molecular techniques. J. Med. Microbiol. 57 1364-1368. [ Links ]
BANDO SY, ANDRADE FB, GUTH BE, ELIAS WP, MOREIRA-FILHO CA and PESTANA DE CASTRO AF (2009) Atypical enteropathogenic Escherichia coli genomic background allows the acquisition of non-EPEC virulence factors. FEMS Microbiol. Lett. 299 22-30. [ Links ]
BOOM R, SOL CJA, SALIMANS MMM, JANSEN CL, WERTHEIM-VAN DILLEN PME and VAN DER NOORDAA J (1990) Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28 495-503. [ Links ]
BOOM R, COL C, BELD M, WEEL J, GOUDSMIT J and VAN DILLEN PW (1999) Improved silica-guanidinium thiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J. Clin. Microbiol. 37 615-619. [ Links ]
BORODINA TA, LEHRACH H and SOLDATOV AV (2003) DNA purification on homemade silica spin-columns. Anal. Biochem. 321 135-137. [ Links ]
BRÖZEL VS and CLOETE TE (1991) Effect of storage time and temperature on the aerobic plate count and on the community structure of two water samples. Water SA 17 289-300. [ Links ]
COLLINS E, GLENNON M, HANLEY S, MURRAY AM, CORMICAN M, SMITH T and MAHER M (2001) Evaluation of a PCR/DNA probe colorimetric membrane assay for identification of Campylobacter spp. in human stool specimens. J. Clin. Microbiol. 39 4163-4165. [ Links ]
DEDEIĆ-LJUBOVIĆ A, HUKIĆ M, BEKIĆ D and ZVIZDIĆ A (2009) Frequency and distribution of diarrhoeagenic Escherichia coli strains isolated from pediatric patients with diarrhoea in Bosnia and Herzegovina. Bosnian J. Basic Med. Sci. 9 148-155. [ Links ]
DUNCKER LC, MATSEBE GN and MOILWA N (2007) The social/cultural acceptability of using human excreta (faeces & urine) for food production in rural settlements in South Africa. WRC Report No. TT 310/07. Water Research Commission, Pretoria, South Africa. [ Links ]
ESTRADA-GARCIA T, LOPEZ-SAUCEDO C, THOMPSON-BONILLA R, ABONCE M, LOPEZ-HERNANDEZ D, SANTOS JI, ROSADO JL, DUPONT HL and LONG KZ (2009) Association of diarrheagenic Escherichia coli pathotypes with infection and diarrhea among Mexican children and association of atypical enteropathogenic E. coli with acute diarrhea. J. Clin. Microbiol. 47 93-98. [ Links ]
FOLTON P, SHEPPARD S, KONVICKA M and SYMONDSON WOC (2005) The significance of facultative scavenging in generalist predator nutrition: detecting decayed prey in the guts of predators using PCR. Mol. Ecol. 14 4147-4158. [ Links ]
GALBADAGE T, JIANG ZD and DUPONT HL (2009) Improvement in detection of enterotoxigenic Escherichia coli in patients with travelers' diarrhea by increasing the number of E. coli colonies tested. Am. J. Trop. Med. Hyg. 80 20-23. [ Links ]
GENTRY-WEEKS C, HUTCHESON HJ, KIM LM, BOLTE D, TRAUB-DARGATZ J, MORLEY P, POWERS B and JESSEN M (2002) Identification of two phylogenetically related organisms from feces by PCR for detection of Salmonella spp. J. Clin. Microbiol. 40 1487-1492. [ Links ]
GUBALA AJ (2006) Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 65 278-293. [ Links ]
HONG HA (2007) Pro-inflammatory Cytokines as Biomarkers of Infection Caused by Human Exposure to Diarrhogenic Escherichia coli. M. Tech Dissertation, University of Johannesburg. [ Links ]
HSU SC and TSEN HY (2001) PCR primers des'igned from malic acid dehydrogenase gene and their use for detection of Escherichia coli in water and milk samples. Int. J. Food Microbiol. 64 1-11. [ Links ]
JAFARI F, GARCIA-GIL LJ, SALMANZADEH-AHRABI S, SHOKRZADEH L, ASLANI MM, POURHOSEINGHOLI MA, DERAKHSHAN F and ZALI MR (2009) Diagnosis and prevalence of enteropathogenic bacteria in children less than 5 years of age with acute diarrhea in Tehran children's hospitals. J. Infect. 58 21-27. [ Links ]
JAGALS P, BARNARD TG and TRAORE HN (2006) Inflammatory potential measurement as a supplement to health-related microbial water-quality assessment. WRC Report No. 1444/1/06. Water Research Commission, Pretoria, South Africa. [ Links ]
KHAN AM, HOSSAIN MS, KHAN AI, CHISTI MJ, CHOWDHURY F, FARUQUE ASG and SALAM MA (2008) Bacterial enteropathogens of neonates admitted to an urban diarrhoeal hospital in Bangladesh. J. Trop. Pediatr. 10 1093. [ Links ]
KONG RYC, LEE SKY, LAW TWF, LAW SHW and WU RSS (2002) Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 36 2802-2812. [ Links ]
MCORIST AL, JACKSON M and BIRD AR (2002) A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50 131-139. [ Links ]
MERAZ IM, JIANG ZD, ERICSSON CD, BOURGEOIS AL, STEFFEN R, TAYLOR DN, HERNANDEZ N and DUPONT HL (2008) Enterotoxigenic Escherichia coli and diffusely adherent E. coli as likely causes of a proportion of pathogen-negative travelers' diarrhea - a PCR based study J. Travel Med. 15 412-418. [ Links ]
NECHVATAL JM, RAM JL, BASSON MD, NAMPRACHAN P, NIEC SR, BADSHA KZ, MATHERLY LH, MAJUMDAR APN and KATO I (2008) Faecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J. Microbiol. Methods 72 124-132. [ Links ]
PATON AW and PATON JC (1998) Detection and characterization of shiga toxigenic Escherichia coli by using multiplex PCR assay for stx1, stx2, eaeA, EHEC hlyA and rfb. J. Clin. Microbiol. 36 598-602. [ Links ]
RAM S, VAJPAYEE P and SHANKER R (2009) Enterotoxigenic Escherichia coli in sewage-impacted waters and aquatic weeds: quantitative PCR for culture-independent enumeration. J. Appl. Microbiol. 108 (3) 1007-1014. [ Links ]
REYES D, VILCHEZ S, PANIAGUA M, COLQUE P, WEINTRAUB A, MOLLBY R and KUHN I (2009) Diversity of intestinal Escherichia coli populations in Nicaraguan children with/ without diarrhoea. J. Med. Microbiol. 58 1593-1600. [ Links ]
SILVA VL, NICOLI JR, NASCIMENTO TC and DINIZ CG (2008) Diarrheagenic Escherichia coli strains recovered from urban pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr. Microbiol. 59 302-308. [ Links ]
STONE GG, OBERST RD, HAYS MP, McVEY S and CHENGAPPA MM (1994) Detection of Salmonella serovars from clinical samples by enrichment broth cultivation–PCR procedure. J. Clin. Microbiol. 32 1742-1749. [ Links ]
TARR CL, PATEL JS, PUHR ND, SOWERS EG, BOPP CA and STROCKBINE NA (2007) Identification of Vibrio isolates by a Multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 45 145-150. [ Links ]
USEIN CR, TATU-CHITOIU D, CIONTEA S, CONDEI M and DAMIAN M (2009) Escherichia coli pathotypes associated with diarrhea in Romanian children younger than 5 years of age. Jap. J. Infect. Dis. 62 289-293. [ Links ]
ZHANG BW, LI M, MA LC and WEI FW (2006) A widely applicable protocol for DNA isolation from fecal samples. Biochem. Genet. 44 503-512. [ Links ]
This paper was originally presented at the 2010 Southern African Young Water Professionals Conference, Pretoria, 19-20 January 2010
* To whom all correspondence should be addressed
+2711 559 6567; fax: +2711 559 6329
e-mail: tgbarnard@uj.ac.za














