Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
Water SA
versão On-line ISSN 1816-7950
versão impressa ISSN 0378-4738
Water SA vol.36 no.1 Pretoria Jan. 2010
Effect of phenolic compounds on the rapid direct enzymatic detection of β-D-galactosidase and β-D-glucuronidase
Sagaran Abboo; Brett I Pletschke*
Department of Biochemistry, Microbiology and Biotechnology, Rhodes University, PO Box 94, Grahamstown 6140, South Africa
ABSTRACT
β-D-Galactosidase and β-D-glucuronidase are 2 marker enzymes used in the rapid detection of total coliforms and E. coli, respectively. A range of bioprobes and biosensors have recently been developed for the rapid, direct and in situ detection of these enzymes. Chromogenic substrates are often used to assay for these enzymes and result in phenolic products being formed. However, phenolic compounds may also be present in water due to industrial activity. In this study, the effect of 11 US EPA priority pollutant phenols (PPP) on these enzyme assays were investigated and it was shown that over- and under-estimation of β-D-galactosidase and β-D-glucuronidase activities may occur due to inhibition or activation of these enzymes in the presence of these phenolic compounds. The types of inhibition as well as inhibition constant (Ki) values were established for the inhibited activities. Wastewater treatment plant and other effluents (e.g. tannery effluents) may contain these phenolic compounds at concentrations high enough to inhibit or activate the activities of the marker enzymes, therefore influencing the rapid and direct enzymatic measurement of faecal contamination using these metabolic marker enzymes.
Keywords: coliforms, E. coli, faecal, β-D-galactosidase, β-D-glucuronidase, inhibition, wastewater.
Introduction
For the detection of indicator bacteria such as total coliforms or Escherichia coli (E. coli), microbial enzyme profiles are a preferred option compared to classical methods, because the reactions are more sensitive and rapid to perform and bacteria can be detected and enumerated through specific enzyme activities (Romprẻ et al., 2002). Chromogenic or fluorogenic enzyme substrates are used to detect the enzymes β-D-galactosidase (β-GAL) and β-D-glucuronidase (β-GUD). The chromogenic enzyme substrates are phenol-based, for example the enzyme substrates for β-GUD are p-nitrophenyl-β-D-glucuronide (PNPG) and 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (XGLU), where PNPG produces ρ-nitrophenol (a yellow colour) and XGLU produces a blue indoxyl product. The fluorogenic substrate used to detect β-GUD is 4-methylumbelliferyl-b- D-glucuronide (MUG) (Manafi, 1996; 2000). β-GAL is detected by using the chromogenic substrates chlorophenol red-β-galactopyranoside (CPRG) and o-nitrophenyl-β-D-galactopyranoside (ONPG) (Edberg et al., 1988; Cheng et al., 2002).
As phenolic compounds have extensive industrial applications, they are often found to be present in the aquatic environment (Llompart et al., 2002; Asan and Isildak, 2003). Several major sources are responsible for the presence of these phenolic compounds in the environment, e.g. pesticides, bactericides, wood preservatives and dyes. Phenolic compounds are also present in pulp processing, petroleum refining, leather tannery, textiles and plastics (Lee et al., 1996; Angelino and Gennaro, 1997; Pẽnalver et al., 2002; Asan and Isildak, 2003; Lupetti et al., 2004). Furthermore, phenols may also be found in fertilizers and explosives (Aktas et al., 2006).
The US Environmental Protection Agency (EPA) lists 11 priority pollutant phenols (PPP), according to their levels of toxicity, which are characterised by the substituent chloro, nitro and methyl groups (Angelino and Gennaro, 1997; Llompart et al., 2002). These 11 PPP compounds are: phenol; 4-nitrophenol; 2-chlorophenol; 2-nitrophenol; 2,4- dinitrophenol; 2,4-dimethylphenol; 4-chloro-3-methylphenol; 4,6-dinitro-2-methylphenol; 2,4-dichlorophenol; 2,4,6-triclorophenol and pentachlorophenol. The European Union (EU) lists the following 7 highly toxic phenols in its legislation: 2-amino-4-chlorophenol; 4-chloro-3-methylphenol; 2-chlorophenol; 3-chlorophenol; 4-chlorophenol; trichlorophenol and pentachlorophenol (Gonzalez-Toledo et al., 2001). The limits for the phenols are set at a maximum of 0.5 ug·ℓ-1 for total phenols and 0.1 µg·ℓ-1 for each individual phenol (Lee at al., 1996).
Nistor et al. (2002) measured the amount of phenolic compounds in environmental water samples at different stages of wastewater treatment. The 3 sources of effluent were at raw, primary and final stages. The concentrations of phenolic compounds were as high as 0.114 mg·mℓ-1 for nitrophenol at the primary effluent stage and 0.083 mg·mℓ-1 for 2,3-dichlorophenol at the raw effluent stage. These results were obtained by solid phase extraction-liquid chromatography-mass spectrometry (SPE-LC-MS) analysis. Based on these findings, the concentrations of phenolic compounds used in this study to examine the effect of interference of these phenolic compounds on the β-GAL and β-GUD assays were set at 0.1 mg·mℓ-1 and 0.2 mg·mℓ-1, respectively.
β-GAL and β-GUD enzymes and their respective activities have been used in the design and development of rapid and direct in situ bioprobe assays and biosensors for faecal detection and monitoring (Wutor et al., 2007a;b;c; Fiksdal and Tryland, 2008). The primary aim of this study was to investigate the potential effect of the PPP compounds at levels commonly found in wastewater on β-GAL and β-GUD activity assays using the substrates PNPG and CPRG.
Experimental
β-D-galactosidase (β-GAL) assays
The CPRG assay performed in this study was adapted from Cheng et al. (2002). The assay buffer used was a 0.1 M sodium phosphate buffer, pH 7.0. A substrate solution of 0.75 mM CPRG (Roche Molecular Biochemicals), diluted in the sodium phosphate buffer, was prepared. A 100 μℓ aliquot stock solution in buffer of commercial β-GAL enzyme (Sigma Aldrich G-2519, EC 3.2.1.23) was then added to a well in a microtitre plate to give a final concentration of 6.88 x 10-5 nM, followed by 100 μℓ of the CPRG solution to initiate the reaction. The change in absorbance was detected at 550 nm at 37ºC for 30 min, reading at 1 min intervals on a Powerwavex microtitre-plate reader (Bio-Tek Instruments, Inc., USA). All assays were performed in triplicate.
β-D-glucuronidase (β-GUD) assays
The enzyme assay used to determine β-GUD activity was adapted from Fisher and Woods (2000). The substrate PNPG was prepared in assay buffer at a concentration of 10 mM. The assay buffer consisted of 0.1 M Tris-HCl, pH 8.0 containing 0.6 mM CaCl2. Commercial β-GUD was obtained from Sigma-Aldrich (G-7396, EC 3.21.31) and prepared in the assay buffer above. A 100 μℓ aliquot stock solution of enzyme in buffer was placed in a microtitre-plate well to give a final concentration of 6.88 x 10-5 nM, after which 100 μℓ of PNPG substrate was added to initiate the reaction which was monitored at 405 nm for 10 min taking readings every 30 s at 37ºC on a Powerwavex microtitre-plate reader (Bio-Tek Instruments, Inc., USA). All assays were performed in triplicate.
β-GAL and β-GUD assays in the presence of phenolic compounds
In order to observe the effects of the 11 PPP compounds listed above on β-GAL and β-GUD enzyme activity, the same assay parameters were used as described above but with the addition of each individual phenolic compound. Individual stock solutions (0.2 mg·mℓ-1) of each phenolic compound were prepared by dissolving the compounds in either 100% acetonitrile or dimethylsulfoxide (DMSO), and then diluting using an appropriate amount of Milli-Q grade water to the required volume. A mixture of the respective enzyme and individual phenolic compound was prepared to give final concentrations of 0.1 mg·mℓ-1 and 0.2 mg·mℓ-1 of each phenolic compound, added to the wells in the microtitre plate, and placed in the Powerwavex microtitre-plate reader until a temperature of 37ºC was reached. The respective substrates were then added and the reactions were monitored. Appropriate controls for the contribution of buffer and enzyme, and substrate and buffer were performed at each phenol concentration used. All assays were performed in triplicate.
Results and discussion
Selection of organic solvents to dissolve the phenolic compounds
Since phenolic compounds are not totally soluble in water, an organic solvent was initially used to dissolve these compounds prior to adding Milli-Q grade water to the desired volume for stock solutions. It was therefore crucial that the organic solvent used to dissolve the phenolic compounds would not interfere with the enzyme assays, so that only the effects of the 11 PPP compounds being monitored would be observed. Acetonitrile was initially selected as the organic solvent for this purpose. Studies were performed to establish whether or not acetonitrile contributed to any background interference. These experiments were performed with both enzyme assays using only the enzyme (β-GAL or β-GUD) and acetonitrile without any of the phenolic compounds present. The organic solvent was tested at the same concentrations as those present during the investigation of the effects of the 11 PPP compounds on the β-GAL and β-GUD assays.
When tested in the β-GUD assay acetonitrile was shown to be a compatible solvent, resulting in little background interference (data not shown). When tested in the β-GAL assay; however, acetonitrile contributed to some background interference, at a concentration of 1% (v/v). For this reason, alternative organic solvents were therefore also investigated. The most compatible solvent which was selected to dissolve the phenolic compounds for the β-GAL assay was 100% dimethylsulfoxide (DMSO)
Effects of 11 PPP compounds on the β-GAL and β-GUD enzyme assays
A concentration of 0.1 mg·mℓ-1 of each of the phenolic compounds was added individually to the β-GAL and β-GUD assays. These were done in triplicate in microtitre 96 well plates and read in the Powerwavex under the above conditions for the respective assays. The blank consisted of buffer and substrate. The positive control consisted of buffer, enzyme and substrate and this represented 100% enzyme activity.
Two compounds (namely 4-nitrophenol and 2,4-dinitrophenol) activated β-GUD enzyme activity, as can be seen in Fig. 1. For both of these compounds the mean value exceeded the maximum absorbance of 4 mOD (milli optical density). Eight of the compounds produced an inhibitory effect (Fig. 1), with the 11th compound (2-nitrophenol) exhibiting a large negative absorbance and interfering with the β-GUD assay (data not shown).
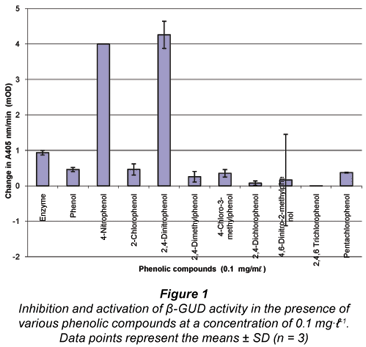
As the compounds 4-nitrophenol and 2,4-dinitrophenol showed a dramatic activation on the β-GUD enzyme assay these were selected for further study. Furthermore, 4-nitrophenol is also the product formed during the β-GUD enzyme assay when the substrate PNPG is added. Three other PPP compounds (2,4-dimethylphenol, 4-chloro-3-methylphenol and 2,4-dichlorophenol) were also selected for further study. The results above were observed when the 11 PPP compounds were added individually at a concentration of 0.1 mg·mℓ-1 to the β-GUD enzyme assay. Statistical analysis revealed that all 5 of the selected phenolic compounds had a significant effect on the activity of β-GUD (P < 0.05 using Microsoft Excel 2003 statistical tool at 5% level of significance).
The results obtained when the 11 PPP compounds were added to the β-GAL assay showed that all the compounds (with the exception of 2-chlorophenol and 2-nitrophenol) had an inhibitory effect on the enzyme assay (see Fig. 2). The compound 2-chlorophenol resulted in a similar absorbance to the positive β-GAL control (i.e. no phenolic compound present – 'Enzyme'). The compound 2,4-dichlorophenol exhibited the greatest degree of inhibition, followed closely by 4,6-dinitro-2-methylphenol; 4-chloro-3-methylphenol and pentachlorophenol.
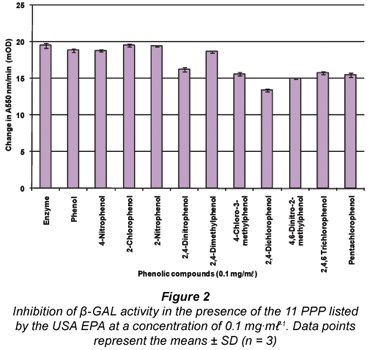
For the β-GAL enzyme assay, 4 of the PPP compounds; namely 4-chloro-3-methylphenol, 2,4-dichlorophenol, 4,6- dinitro-2-methylphenol and pentachlorophenol, were selected for further study on the basis that these compounds exhibited the highest degree of inhibition to the activity of the β-GAL enzyme assay when added. This selection process was based on the results in Fig. 2 representing the effects of the 11 PPP compounds added at a concentration of 0.1 mg·mℓ-1 to the β-GAL enzyme assay. Statistical analysis revealed that the effects of these phenols were indeed significant. A 5th compound (2-chlorophenol) was also selected based on the fact that it is similar to chlorophenol red, which is the product formed in the β-GAL enzyme assay when the substrate CPRG is hydrolysed. However, the effect of 2-chlorophenol was not significant (P>0.05).
Types of inhibition and inhibition constants (Ki values)
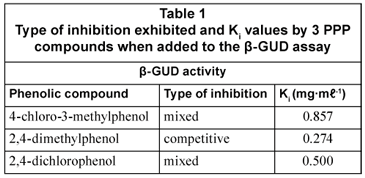
Detailed investigations were performed on each of the five selected compounds for β-GAL and β-GUD. These were in the form of inhibition studies performed to test the interference of the selected phenolic compounds on the β-GUD and β-GAL assays by investigating the effects of 0.1 mg·mℓ-1 and 0.2 mg·mℓ-1 of each phenolic compound on the enzyme assays at various levels of substrate varying from 0 - 7.937 mM (for PNPG) and 0 to 1.645 mM (for CPRG). Double-reciprocal Lineweaver-Burk plots for each phenolic compound were then constructed using the data obtained. From these graphs, the type of inhibition was elucidated (Wilson and Walker, 2005). Three types of inhibition were identified; competitive, non-competitive and mixed inhibition. Secondary plots were then constructed in order for the inhibition constant Ki to be calculated (Wilson and Walker, 2005).
Both 4-chloro-3-methylphenol and 2,4-dichlorophenol demonstrated mixed inhibition in the β -GUD assay; while 2,4-dimethylphenol showed competitive inhibition (Table 1). The inhibitory effect on the β-GUD enzyme assay was attributed to 2,4-dimethylphenol competing for the same binding site as the substrate PNPG on the β-GUD molecule. The compounds 4-chloro-3-methylphenol and 2,4-dichlorophenol bound to the same active site as the substrate PNPG as well as an alternate binding site on the β-GUD enzyme molecule.
In the β-GAL enzyme assay, the types of inhibition exhibited by 4 of the PPP compounds tested were as follows: 4-chloro-3-methylphenol (competitive); 4,6-dinitro-2-methylphenol (competitive); 2,4-dichlorophenol (non-competitive) and pentachlorophenol (competitive). The 5th phenolic compound, 2-chlorophenol, exhibited no significant effect on the normal reaction observed in the absence of inhibitor. These results are listed in Table 2. The inhibition described above was attributed to 4-chloro-3-methylphenol, 4,6-dinitro-2-methylphenol and pentachlorophenol competing for the same binding site as the substrate CPRG, since this study revealed competitive inhibition. The compound 2,4-dichlorophenol also bound to an alternative binding site on the β-GAL enzyme molecule, as in the case of the non-competitive inhibition. This resulted in a decrease in velocity of the enzyme reaction.
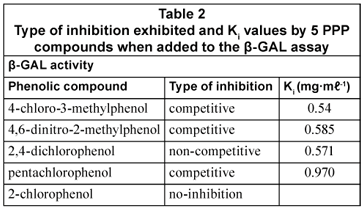
The inhibition constants (Kis) of the PPP compounds for the 2 enzymes were generally of the same order of magnitude, indicating that these PPP compounds generally had approximately the same affinities for the 2 enzymes.
Activation of the β-GUD enzyme assay
Figure 3 illustrates the activation of the β-GUD enzyme assay with increasing concentrations of 4-nitrophenol and 2,4-dinitrophenol. At 100 µg/mℓ addition 4-nitrophenol exceeded the maximum absorbance.
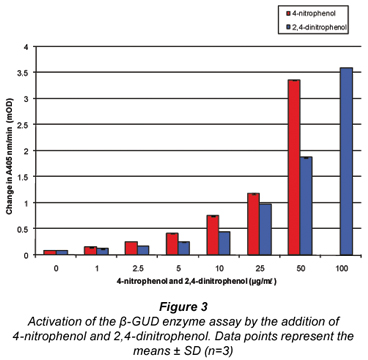
This activation occurred due to the fact that the product formed in the β-GUD enzyme assay was also 4-nitrophenol, hence the activation when 4-nitrophenol and 2,4-dinitrophenol were added to the assay. These phenolic compounds are either identical (4-nitrophenol) or similar (2,4-dinitrophenol) to the product (4-nitrophenol) formed in the β-GUD enzyme assay.
Several of the 11 phenolic compounds listed by the US EPA present in the environment were therefore indeed able to affect β-GAL and β-GUD enzyme activity. There is therefore a possibility that an over- or under-estimation of β-GAL and β-GUD enzyme activities can result when these phenolic compounds are present in the environment.
The different PPP compounds clearly had varied effects on the activities of both enzymes. These effects were also concentration-dependent. These phenolic compounds competed for the same active sites as the substrates used in these assays as well as combined to alternate sites on the enzyme molecules. In mixed inhibition the inhibitors could also be binding to the enzyme-substrate complex, thereby reducing the overall velocity of the reaction.
Conclusions
Several of the 11 PPP compounds listed by the US EPA in the environment were able to affect β-GAL and β-GUD enzyme activities at levels of 0.1 mg·mℓ-1 or greater. Environmental samples, e.g. wastewater treatment plant (WWTP) effluents do contain these phenolic compounds at these levels (Nistor et al., 2002). Nistor et al. (2002) reported a concentration of 0.114 mg·mℓ-1 for nitrophenol at the primary effluent stage and 0.083 mg·mℓ-1 for 2,3-dichlorophenol at the raw effluent stage. Therefore, when rapid and direct enzymatic measurement of faecal contamination is performed using these metabolic marker enzymes in effluents where high levels of PPP compounds are thought to be present, e.g. in WWTP effluents, the results may be affected by these compounds. Results using these rapid and direct enzyme assays can and should be further verified by using more traditional microbiological water quality methods for the detection of coliforms or E. coli. The rapid and direct enzyme assay methods, however, still serve as a rapid and effective early detection method for detecting the potential presence of faecal contamination in water for human consumption or recreational use.
Acknowledgements
This material is based upon work supported by the National Research Foundation (NRF) and the Water Research Commission (WRC) of South Africa. Any opinion, findings and conclusions or recommendations expressed in this material are those of the author(s) and therefore the NRF does not accept any liability in regard thereto.
References
AKTAS AH, SANLI N and PEKCAN G (2006) Spectrometric determination of pKa values for some phenolic compounds in acetonitrile-water mixture. Acta Chim. Slov. 53 214-218. [ Links ]
ANGELINO S and GENNARO MC (1997) An ion-interaction RP-HPLC method for the determination of the eleven EPA priority pollutant phenols. Anal. Chim. Acta 346 61-71. [ Links ]
ASAN A and ISILDAK I (2003) Determination of major phenolic compounds in water by reversed-phase liquid chromatography after pre-column derivatization with benzoyl chloride. J. Chromatogr. A 988 145-149. [ Links ]
CHENG MG, MUNOZ MG, ZHOU H and JIN H (2002) Expression of β-galactosidase by recombinant respiratory syncytial viruses for microneutralization assay. J. Virol. Methods 105 287-296. [ Links ]
EDBERG SC, ALLEN MJ, SMITH DB, LECHEVALLIER M, KRIZ N, CALLAN D, WARD R, CALVER D, JACKSON W, URYC M, STORMS C, LORINER J, TROK T and BURNS M (1988) National field evaluation of a defined substrate method for the simultaneous enumeration of total coliforms and Escherichia coli from drinking water: comparison with the standard multiple tube fermentation method. Appl. Environ. Microbiol. 54 1595-1601. [ Links ]
FIKSDAL L and TRYLAND I (2008) Application of rapid enzyme assay techniques for monitoring of microbial water quality. Curr. Opin. Biotechnol. 19 289-294. [ Links ]
FISHER KL and WOODS JP (2000) Determination of β-glucosidase enzymatic function of the Histoplasma capsulatum H antigen using a native expression system. Gene 247 191-197. [ Links ]
GONZALEZ-TOLEDO E, PRAT MD and ALPENDURADA MF (2001) Solid-phase microextraction coupled to liquid chromatography for the analysis of phenolic compounds in water. J. Chromatogr. A 923 45-52. [ Links ]
LEE SG, HONG SP and SUNG MH (1996) Removal and bioconversion of phenol in wastewater by a thermostable β-tyrosinase. Enzyme Microb. Technol. 19 374-377. [ Links ]
LLOMPART M, LOURIDO M, LANDIN P, GARCIA-JARES C and CELO R (2002) Optimization of a derivatization-solid-phase microextraction method for the analysis of thirty phenolic pollutants in water samples. J. Chromatogr. A 963 137-148. [ Links ]
LUPETTI KO, ROCHA FRP and FATIBELLO-FILHO O (2004) An improved flow system for phenols determination exploiting multicommutation and long pathlength spectrophotometry. Talanta 62 463-467. [ Links ]
MANAFI M (1996) Fluorogenic and chromogenic enzyme substrates in culture media and identification tests. Int. J. Food Microbiol. 31 45-58. [ Links ]
MANAFI M (2000) New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol.60 205-218. [ Links ]
NISTOR C, ROSE A, FARRE M, STOICA L, WOLLENBERGER U, RUZGAS T, PFEIFFER D, BARCELO D, GORTON L and EMNEUS J (2002) In-field monitoring of cleaning efficiency in waste water treatment plants using two phenol-sensitive biosensors. Anal. Chim. Acta 456 3-17. [ Links ]
PEŇALVER A, POCURULL E, BORRULL F and MARCÉ RM (2002) Solid-phase microextraction coupled to high-performance liquid chromatography to determine phenolic compounds in water samples. J. Chromatogr. A 953 79-87. [ Links ]
ROMPRÉ A, SERVAIS P, BAUDART J, DE-ROUBIN MR and LAURENT P (2002) Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49 31-54. [ Links ]
WILSON K and WALKER J (2005) Principles and Techniques of Biochemistry and Molecular Biology (6th edn.). Cambridge University Press, New York. [ Links ]
WUTOR VC, TOGO CA, LIMSON JL and PLETSCHKE BI (2007a) A novel biosensor for the detection and monitoring of β-D-galactosidase of faecal origin in water. Enzyme Microb. Technol. 40 1512-1517. [ Links ]
WUTOR VC, TOGO CA and PLETSCHKE BI (2007b) The effect of physico-chemical parameters and chemical compounds on the activity of β-d-galactosidase (B-GAL), a marker enzyme for indicator microorganisms in water. Chemosphere 68 622-627. [ Links ]
WUTOR VC, TOGO CA and PLETSCHKE BI (2007c) Comparison of the direct enzyme assay method with the membrane filtration technique in the quantification and monitoring of microbial indicator organisms – seasonal variations in the activities of coliforms and E. coli, temperature and pH. Water SA 33 (1) 107-110. [ Links ]
Received 15 May 2009; accepted in revised form 25 November 2009.
* To whom all correspondence should be addressed.
+2746 6038081; fax: +2746 6223984;
e-mail: B.Pletschke@ru.ac.za














