Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Water SA
On-line version ISSN 1816-7950
Print version ISSN 0378-4738
Water SA vol.36 n.1 Pretoria Jan. 2010
Monitoring bacterial faecal contamination in waters using multiplex real-time PCR assay for Bacteroides spp. and faecal enterococci
RM AgudeloI, II,* F CodonyI; B AdradosI; M FittipaldiI; G PeñuelaII; J MoratóII
ILaboratori de Microbiología Sanitaria i Mediambiental (MSM-Lab) & Aquasost – UNESCO chair in Sustainability, Universitat Politècnica de Catalunya (UPC), Violinista Vellsolá 37, Terrassa – 08222 (Barcelona), Spain
IIGrupo de Diagnóstico y Control de la Contaminación GDCON, Sede de Investigación Universitaria, Universidad de Antioquia, calle 62 # 52-59, Medellin, Colombia
ABSTRACT
Monitoring of sanitary quality or faecal pollution in water is currently based on quantifying some bacterial indicators such as Escherichia coli and faecal enterococci. Using a multiplex real-time PCR assay for faecal enterococci and Bacteroides spp., the detection of faecal contamination in non-treated water can be done in a few hours, reducing the analysis time to 2 h.
The conventional method based on cultures was compared with a multiplex assay procedure for Bacteroides spp. and faecal enterococci with an internal inhibition control. Out of 74 water samples from different sources analyzed, using both procedures, 54 were true positives and 6 true negatives, 12 samples were real-time PCR positive and culture-negative whereas 2 were real-time PCR negative and culture-positive. In conclusion, 89.2% of the samples were found to be positive with real-time PCR and 75.7% with plate cultures.
Detection levels were much higher when using the multiplex real-time PCR assay, based on the higher number of positive samples in comparison with conventional microbiology. The feasibility of multiple reactions in the monitoring of faecal contamination has been demonstrated along with fast quantification of the faecal load. Such procedure can be performed in less than 3 h.
This work extends the use of multiplex real-time PCR for environmental analysis, demonstrating the feasibility of these procedures in monitoring faecal pollution of water samples.
Keywords: faecal contamination, multiplex real-time PCR, water monitoring, faecal enterococci, Bacteroides spp.
Introduction
Pathogenic microorganisms found in non-treated wastewater have the ability to reproduce easily due to the large amount of available nutrients, thereby affecting the environment and presenting a great risk to health (Pusch et al., 2005; Gilbride et al., 2006). Over the past few years, the application of different methods to monitor faecal pollution in diverse water sources has become very important; however, there is no universal approach which fits all requirements to allow completely reliable faecal source identification (Stricker et al., 2008). Currently microbial water quality is evaluated by monitoring the level of a reduced number of microorganism indicators with the use of conventional microbiological procedures based on plate cultures (Tajima et al., 2007). Until now, and despite their clear limitations, the current procedures have been widely used as an operative tool to ensure safe drinking water (Ashbolt et al., 2001). In fact, it is well known that the classical procedures usually fail in the detection of several pathogens, and for this reason in some cases these culture-dependent methods are considered to be outdated (Rose, 2006). Consequently, new tools and new procedures are needed.
Over the past decade, real-time polymerase chain reaction (PCR) methods have become the main alternative to detect the presence of all kinds of microbial pathogens through the amplification of specific DNA sequences and without the need to culture any bacteria (Gilbride et al., 2006). Real-time PCR is considered highly sensitive for the quantification of microorganisms in environmental samples (Ben-Dov et al., 2007), herpes virus in clinical samples (Rose et al., 2008) and intracellular microorganisms in humans and birds, such as Chlamydophila psittaci (Branley et al., 2008).
A PCR laboratory requires a substantial investment compared to conventional microbiology-testing laboratories that use time-consuming and laborious procedures. In our opinion, though, the main obstacles in the establishment of this new technique are the unreliability in differentiating between live or dead microorganisms and the precise quantification of the targets. Lately this situation has been changing due to the evolution of conventional PCR to real-time PCR; the latter technique has been successfully introduced into the environmental field to indentify, quantify and diagnose pathogens and viruses in water faster and with higher sensitivity (Singh and Singh, 2008), including those which cannot be cultivated in cellular lines in an efficient way (Costafreda et al., 2006).
Nevertheless, a lot of work is still required before general consensus is reached that will drive the effort onto different levels of selection, e.g. the choice of best primers and probes for each target, the optimization of multiple reactions to allow for simultaneous detection of different microorganisms or the development of prospective surveys to evaluate the correlation of data between conventional and molecular methods. Multiplex PCR has been used in many investigations with varying objectives, but where similar procedures have been necessary to establish a method to identify and quantify various types of microorganisms, like the production of the granulovirus (GV) isolated from Epinotia aporema (Lepidoptera: Tortricidae), among others (Manzán et al., 2008).
Microbial indicators are usually selected according to several premises, one of which is analytical simplicity. When a process indicator, faecal indicator or model organisms could not be isolated with ease, it would not be considered as a good evaluation tool. Real-time PCR currently offers the possibility to detect and to directly quantify the level of most pathogenic microbes. This is a radical change and as a consequence the list of microbes suitable for use as indicators or models is increasing. Three real-time PCR assays have recently been developed for each coliphage genogroup to discriminate between animal and human sources of faecal contamination (Jones et al., 2009).
In any given case of having to choose a microorganism as a new indicator, real-time PCR can be considered to be the fastest analytical approach. In this study 2 simultaneous detection approaches were evaluated for faecal enterococci and Bacteroides spp. by multiplex real-time PCR in non-treated water samples; the results were compared with conventional microbiological methods. The 1st one, faecal enterococci, is a classical indicator that gives results by conventional procedures within 48 h. The 2nd one, Bacteroides spp., is an interesting indicator of faecal pollution (Savichtcheva and Okabe, 2006), which is not normally used for routine analysis because a simple and fast culture method has not yet been developed.
Materials and methods
Sample selection. All samples were collected from urban areas around Barcelona city (Catalonia, NE of Spain), with multiple sources such as rivers, wells, urban groundwater and wastewater. In total 74 non-treated water samples were taken on different days. The main criterion in the selection of samples was the possible faecal pollution, and therefore potable water sources were not considered for this study.
Conventional microbiology assays. Water samples were aseptically collected according to Standard Methods (2000), in Pyrex bottles. Total coliforms and Escherichia coli (E. coli) were enumerated by membrane filtration using Chromocult Coliform Agar (Merck) supplemented with Cefsulodin (Merck), and incubated at 37ºC for 24 h. Faecal enterococci were also enumerated by membrane filtration using Stanlez-Barley Agar (Merck), incubated at 37ºC for 48 h, followed by the confirmation of aesculin hydrolysis by placing the filter into Bile Esculin Agar (REACI00082XA Merck) for 2 h at 44°C. Bacterial counts were expressed as log cfu/100 mℓ.
Sample preparation for real-time PCR. A sample aliquot of 100 mℓ of water from each source was concentrated by membrane filtration using a nylon membrane (0.45 mm porous diameters, Millipore). Cells were re-suspended in 5 mℓ sterile saline solution, by vigorous vortexing for 60 s with 15 glass beads (5 mm diameter) and sonication (JP Selecta S.A., 6 ℓ, 150 W) for 3 min. The cell suspension (4 mℓ) was concentrated to 200 mℓ by centrifugation (Medifriger, JP Selecta S.A.) at 14 000 r/min for 5 min and DNA was extracted with DNeasy Tissue Kit (Qiagen) according to the manufacturer's instructions.
Primers and probes. Three sets of primers and Taqman probes were used. Total Bacteroides spp. according to Layton et al. (2006), with a probe labelled with FAM. Enterococcus faecalis according to He and Jiang (2005), with a probe labelled with Cy3 dye. An internal inhibition control was used, a human beta-actin according to Kalina et al. (1997), and labelled with Cy5 dye. Sequences of the oligonucleotides are shown in Table 1.
Real-time assays. An aliquot of 5 mℓ of DNA plus 20 mℓ of master mix was analysed in duplicate and the amplification was performed on a Cepheid Smartcycler 2.0, which protocol is presented in Table 2. The master mix included 10 mℓ Quantitec Multiplex PCR mix, Qiagen; 0.5 U/reaction of uracil-DNA glycosylase (UDG) (New England Biolabs) and 5 pg/μℓ of human DNA (Thermo scientific) as a target for the internal inhibition control. The sequences of oligonucleotides (primers and probes) were acquired from the firm Thermo, at a concentration of 50 µM. For the PCR, the procedures were followed as instructed by the supplier.
Bacterial standards. Overnight cultures (Bacteroides spp. ATCC 51477, Enterococcus faecalis NCTC 775) plated in brain heart infusion agar (BHIA) (Merck), and stored at 37°C for 3 d, were used to prepare the standard bacterial suspensions. Turbidity was optically measured with a spectrophotometer and adjusted to 0.2 absorbance units at 600 nm, corresponding to approximately 1·108 cfu/mℓ. Colony-forming units were also determined by plating on BHIA agar (Merck). Bacteroides were anaerobically cultured using the anaerocult system (Merck).
Multiplex PCR efficiency evaluation. Serial logarithmic dilutions, from 1·101 to 1·105 cfu/reactions, of each DNA standard, were performed and mixed. The combination of both standards was made in an inverse order, in the way that the highest concentration of Bacteroides spp. DNA was mixed with the lowest concentration of Enterococcus faecalis DNA, and vice versa. The amplification efficiency was calculated according to estimation by means of the slope calculation method from a calibration dilution curve (Rasmussen, 2001).
Results
The multiplex real-time PCR method efficiency for faecal enterococci and Bacteroides spp. was, respectively, 1.93 and 2.11. For both cases the value is near 2.0 which is the theoretical optimum number (2.0 corresponds to 100% base-10 logarithmic scale). The multiplex reactions ran without any cross interferences between the different primers and probes.
Seventy-four non-treated water samples from different sources were analysed. In any one of the tested samples, the PCR inhibition was detected with an internal control. Table 3 shows the comparison of qualitative results. The sample was considered positive for faecal pollution by culture or PCR methods, even when only 1 of the 2 targets was detected (faecal enterococci-E. coli by culture or faecal enterococci-Bacteroides by real-time PCR). With this approach, 54 samples were true positive and 6 true negative by both methods, 12 samples were PCR positive and culture negative, whereas 2 samples were culture positive and PCR negative. In conclusion, 89.2% of the samples were positive with real-time PCR and 75.7% with plate cultures. According to the normative, ISO/TR 13843 (2000) and ICONTEC GTC/84 (2003), the relative accuracy of real-time method in relation to conventional methods, was high, having an efficiency of 81.1% and a sensitivity of 96.4%.
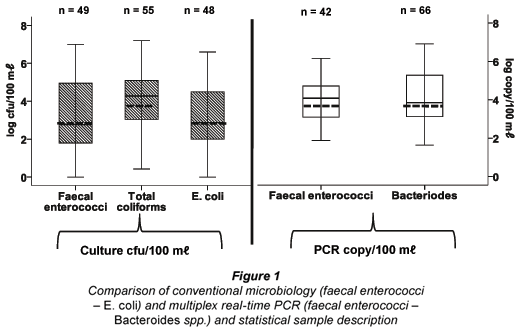
The quantitative results given in Fig. 1 show that the bacterial quantification data were more homogeneous when using PCR than when using conventional culture microbiology. Most of the samples were positive with values ranging between 0 and 7 log units, depending on the water type and the monitoring method. Real-time PCR was the method which gave the highest number of detected positive samples for Bacteroides (n = 66) and the conventional culture method gave the highest number of positive samples for total coliforms (n = 55). It is also shown in Fig. 1 that the average higher concentration for total coliform (4.11 cfu/100 mℓ) was obtained by culturing compared with the other 2 bacterial types that were analyzed. With multiplex real-time PCR, the average quantity of faecal enterococci was higher than that found by culture.
To evaluate the contamination level and to compare conventional with molecular methods, the highest value of each pair (faecal enterococci-E. coli or faecal enterococci-Bacteroides) was selected. The correlation of positive values for conventional microbiology and real-time PCR (Fig. 2) showed a positive correlation and a coefficient of correlation of 0.74. The counts by real-time PCR were 1.1 log units (with a confidence interval of 95.0%; 0.73 to 1.41) higher than those obtained by conventional culture-method microbiology.

Discussion
In this study 2 different analytical strategies were used for the evaluation of faecal presence in non-treated water. With conventional methods based on biochemical characteristics of colonies growing in selective and differential culture media, faecal enterococci and E. coli were studied and compared with molecular methods that recognize specific sequences of the bacterial genome. Two different targets were used with the molecular approach: a classical indicator (faecal enterococci) and an alternative one (Bacteroides spp.). Although both strategies had identical purposes and objectives, the results and the units were found to be different.
To ensure equivalence between the 2 analytical methods, a qualitative comparison based on the ISO/TR 13843 (2000) could be an appropriate approach. However, it has to be taken into account that this involves the use of 2 different concepts: colony-forming units vs. genetic sequence copies and that the accuracy and sensitivity of the results were not close to 100.0%. When 2 quantitative methods are compared, however, there are some aspects of variability in micro-organisms that have to be taken into account. The microorganisms are not solutes like ions, which for chemical analyses can be assumed to be homogeneously distributed. When introduced into water, microorganisms do not form a perfect solution but a suspension, which imparts a degree of significance on inherent heterogeneity (Sartory, 2005).
On the other hand the correlation in the quantitative detection of faecal pollution is indeed relatively good. Combining 2 different bacterial targets in a multiplex reaction, the agreement with conventional culture-based methods presented a correlation coefficient of 0.74. Similar results and correlations are found in other works, e.g., the connection between E. coli by culture and Bacteroides rumiant specific markers showed correlation coefficients of between 0.72 and 0.80 (Reischer et al., 2008).
In this study, another important dispersion factor was found: the 2 different pairs of compared bacterial species. It is obvious that the pair of bacterial species used for real-time PCR (Bacteroides spp. and faecal enterococci) is not the same or even equivalent to the pair used for the method by culture (E. coli and faecal enterococci). In this study, efforts were aimed at comparing faecal detection using conventional and molecular approaches, and disregarding the different target bacteria.
The real-time PCR assay is a more appropriate method to detect positive samples than plate-culturing methods. When both methods are compared in a general way, the molecular approach proved to more sensitive and detected lower levels of faecal pollution in most cases. The amount of contamination detected by real-time PCR is no less than 1 log higher than the amount detected by culture. This difference between plate counts and molecular detection can be explained by the presence of viable but non-culturable bacteria (VBNC) (Olivier, 2000; Szewzyk et al., 2000).
There may be another difference in results when viable plate counts do not detect dead cells that may be positively detected by PCR (Josephson et al., 1993). Although some results using Legionella pneumophila (Bej et al., 1991) have shown that VBNC bacterial cells showed positive amplification in opposition with the non-viable cells, this remains unclear in the field of water microbiology, and to date controversy remains. For example, Shih and Lin, 2006 postulate that the false-positive results encountered by Joly et al. (2006) can be explained by the presence of nonviable Legionella cells in water samples.
Combining microbial culture or real-time PCR methods, allows obtaining a wider range of results in pollution evaluation. Both methods are based on different approaches: the first detects vegetative and cultivable cells and the latter detects genetic targets; for this reason it is not surprising that a perfect equivalence between them does not exist. As stated in Table 2, molecular methods are capable of detecting more polluted samples; nevertheless, there are 2 samples in which real-time PCR fails, but these results are explained with a multiple approach. It is well known that quantification by molecular methods obtains, in the absence of PCR inhibition, higher counts than culture methods. Monitoring of genetic targets also detects dead cells, viable but non-cultivable cells (Hussong et al., 1987), and counts all the individual cells which form aggregates. It is for this reason that, in most cases, molecular methods are more sensitive than culture methods.
Over the past number of years, publication of viable PCR methods which are based on the use of propidium monoazide (PMA) or ethidium monoazide (EMA) (Nogva et al., 2003; Nocker et al., 2006) demonstrated the feasibility of detecting only viable cells in assays with different bacteria (Rudi et al., 2005; Soejima et al., 2007; Cawthorn and Witthuhn, 2008; Bae and Wuertz, 2009), spores (Rawsthorne et al., 2009) and fungi (Vesper et al., 2008). Although there are interesting publications, consensus on methodology still has not been reached. In our opinion, this future scenario will improve the correlation between culture and molecular methods. On the other hand, although there is a growing tendency to consider more sensitive molecular methods rather than culture ones, in some cases that is not completely effective. For example, for a membrane filtration method, the complete sample volume is cultured because the filter is placed over the culture media, while for molecular analysis several steps like elution, concentration and nucleic acid purification are needed. All these steps do not have a 100% yield, and finally the PCR tube does not contain all the DNA present in the entire sample volume before the treatment steps. For that reason, in samples with a very low level of free, viable and cultivable cells, the culture still will be more sensitive than molecular analysis.
Finally, aggregation is the other factor that needs to be taken into account. While different cells in aggregation might form a unique colony, using real-time PCR each cell from the aggregate is detected in contrast to what occurs in plate-culture methods, in which cell aggregates count only as a colony.
Despite the development of new analytical techniques, the differences between molecular and conventional analysis will always remain, and the future implementation of real-time PCR methods for routine monitoring of faecal pollution in non-treated water will be possible through a better understanding of each result, and specifically their correlation with the associated public health risks.
Conclusions
This study has demonstrated the feasibility of multiple reactions for the monitoring of faecal contamination. Water quality is evaluated with the combinations of several bacterial markers through different microbiological analysis, and the molecular approach, with a single multiplex reaction, uses a combination of different targets, which probably makes it a better evaluation. Also the correlation between culture and real-time PCR in monitoring faecal levels may be improved with multiplex reactions.
The fact that the quantitative real-time PCR approach produces a direct result within a few hours has a more important impact when the environment is endangered and public health is at stake (within 3 h the faecal load in a water sample is known). This method is fast, sensitive and useful to quantify several bacterial types in the same reaction tube along with real-time PCR specificity, and widespread implementation of its protocols should be considered as a promising tool in water quality monitoring.
Acknowledgements
This research was supported by the Project TECSPAR: Sustainability Technologies to the Potabilization and Treatment of Wastewater funded for UE- Programa ALFA. RED ALFA II-0543-FI-FA-FCD; the FPI grant from Ministry of Education and Science of Spain; FI grant from the Comissionat per a Universitats i Recerca del Departament d'Innovació, Universitats i Empresa de la Generalitat de Catalunya i del Fons Social, and the National Faculty of Public Health, University of Antioquia (Colombia). Aigües de Terrassa Company is gratefully acknowledged for supplying water samples.
References
ASHBOLT NJ, GRABOW WOK and SNOZZI M (2001) Indicators of microbial water quality. In: Fewtrell L and Bartram J (eds.) Water Quality: Guidelines, Standards and Health. Assessment of Risk and Risk Management for Water Related Infectious Disease. WHO Water Series. IWA Publishing, London. 285-315. [ Links ]
BAE S and WUERTZ S (2009) Discrimination of Viable and Dead Fecal Bacteroidales Bacteria by Quantitative PCR with Propidium Monoazide. Appl. Environ. Microbiol. 75 2940-2944. [ Links ]
BEJ AK, MAHBUBANI MH and ATLAS RM (1991) Detection of viable Legionella pneumophila in water by polymerase chain reaction and gene probe methods. Appl. Environ. Microbiol. 57 (2) 597-600. [ Links ]
BEN-DOV E, BRENNER A and KUSHMARO A (2007) Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb. Ecol. 54 (3) 439-451. [ Links ]
BRANLEY JM, ROY B, DWYER DE and SORREL TC (2008) Real-time PCR detection and quantitation of Chlamydophila psittaci in human and avian specimens from a veterinary clinic cluster. Eur. J. Clin. Microbiol. Infect. Dis. 27 269-273. [ Links ]
CAWTHORN DM and WITTHUHN RC (2008) Selective PCR detection of viable Enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J. Appl. Microbiol. 105 1178-1185. [ Links ]
COSTAFREDA M, BOSCH A and PINTO R (2006) Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay, for quantification of Hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72 (6) 3846-3855. [ Links ]
GILBRIDE KA, LEE DY and BEAUDETTE LA (2006) Molecular techniques in wastewater: understanding microbial communities, detecting pathogens, and real-time process control. J. Microbiol. Method 66 1-20. [ Links ]
HE JW and JIANG S (2005) Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71 (5) 2250-2255. [ Links ]
HUSSONG D, COLWELL RR, O'BRIEN MO, WEISS E, PEARSON AD, WEINER RM and BURGE WD (1987) Viable Legionella pneumophila not detectable by culture on agar media. Biotechnol. 5 947-950. [ Links ]
ISO (2000) Water Quality – Guidance on Validation of Microbiological Methods. ISO/TR 13843:2000. Geneva: International Organization for Standardization. [ Links ]
ICONTEC (2003) Calidad del agua Guía para la orientación acerca de la validación de métodos de análisis microbiológicos. GTC/ 84. Bogotá: Instituto Colombiano de Normas Técnicas y Certificación. [ Links ]
JOLY P, FALCONNET PA, ANDRÉ J, WEILL N, REYROLLE M, VANDENESCH F, MAURIN M and JARRAUD S (2006) Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl. Environ. Microbiol. 72 (4) 2801-2808. [ Links ]
JONES TH, HOUDE A, POITRA E, WARD P and JOHNS MW (2009) Development and evaluation of o Multiplexed Real-time TaqMan RT-PCR assay with a sample process control for detection of F-specific RNA coliphage genogroups I and IV. Food Environ. Virol. DOI 10.1007/s12560-009-9008-7. [ Links ]
JOSEPHSON KL, GERBA CP and PEPPER IL (1993) Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59 3513-3515. [ Links ]
KALINA O, LEBEDEVA I, BROWN J and SILVER J (1997) Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res. 25 (10) 1999-2004. [ Links ]
LAYTON A, McKAY L, WILLIAMS D, GARRETT VR and SAYLER G (2006) Development of bacteroides 16S rRNA gene taqman-based real-time PCR assays for estimation of total, human and bovine faecal pollution in water. Appl. Environ. Microbiol. 72 (6) 4214-4224. [ Links ]
MANZÁN MA, ALJINOVIC EM, BIEDMA ME, SCIOCCO-CAP A, GHIRINGHELLI PD and ROMANOWISKI V (2008) Multiplex PCR and quality control of Epinotia aporema granulovirus production. Virus Genes 37 (2) 203-211. [ Links ]
NOCKER A, CHEUNG CY and CAMPER AK (2006) Comparison of propidium monoazide and ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67 310-320. [ Links ]
NOGVA HK, DROMTORP SM, NISSEN H, and RUDI K (2003) Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease. PCR. Biotech. 810 812-813. [ Links ]
OLIVIER JD (2000) The public health significance of viable but non-culturable bacteria. In: Colwell RR and Grimes DJ (eds.) Nonculturable Microorganisms in the Environment. ASM Press, Washington DC. 277-300. [ Links ]
PUSCH D, OH DY, WOLF S, DUMKE R, SCHRÖTER-BOBSIN U, HÖNE M, RÖSKE I and SCHREIER E (2005) Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 150 929-947. [ Links ]
RASMUSSEN R (2001) Quantification on the light cycler. In: Meuer S, Witwe C and Nakagawara K (eds.) Rapid Cycler Real-Time PCR, Methods and Applications. Springer Press, New York. 21-34. [ Links ]
RAWSTHORNE H, DOCK CN and JAYKUS LA (2009) PCR-Based method using propidium monoazide to distinguish viable from non viable Bacillus subtilis spores. Appl. Environ. Microbiol. 75 2936-2939. [ Links ]
REISCHER GF, HAIDER JM, SOMME R, STADLER H, KEIBLINGER KM, HORNEK R, ZEROBIN W, MACH RL and FARNLEITNER AH (2008) Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ. Microbiol. 10 2598-2608. [ Links ]
ROSE JB (2006) Identification and characterization of biological risks for water. In: Management of Intentional and Accidental Water Pollution. Nato Security through Science Series. Springer, Netherlands. 85-101. [ Links ]
ROSE L, HERRA CM and CROWLEY B (2008) Evaluation of real-time polymerase chain reaction assays for the detection of herpes simplex virus in swab specimens. Eur. J. Clin. Microbiol. Infect. Dis. 27 827-861. [ Links ]
RUDI K, MOEN B, DRØMTORP SM and HOLCK L (2005) Use of Ethidium Monoazide and PCR in Combination for Quantification of Viable and Dead Cells in Complex Samples. Appl. Environ. Microbiol. 71 1018-1024. [ Links ]
SARTORY DP (2005) Validation, verification and comparison: adopting new methods in water microbiology. Water SA 31 (3) 393-396. [ Links ]
SAVICHTCHEVA O and OKABE S (2006) Alternative indicators of faecal pollution: Relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res. 40 2463-2476. [ Links ]
SHIH HY and LIN YE (2006) Letter to the Editor. Caution on interpretation of Legionella results obtained using real-time PCR for environmental water samples. Appl. Environ. Microbiol. 72 (10) 6859. [ Links ]
SINGH S and SINGH S (2008) Real-time polymerase chain reaction. A revolution in diagnosis. Resonance. 13 (4) 369-377. [ Links ]
SOEJIMA T, IIDA K, QIN T, TANIAI H, SEKI M, TAKADE A and YOSHIDA S (2007) Photo activated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiol. Immunol. 51 763-775. [ Links ]
STANDARD METHODS (2005) Standard Methods for the Examination of Water and Wastewater (Eaton AD, Clesceri LS, Rice EW and Greenberg AE) (21st edn.). United Book Press, USA. 9-111 – 9-136. [ Links ]
STRICKER AR, WILHARTITZ I, FARNLEITNER AH and MACH RL (2008) Development of a Scorpion probe-based real-time PCR for the sensitive quantification of Bacteroides sp. ribosomal DNA from human and cattle origin and evaluation in spring water matrices. Microbiol. Res. 163 (2) 140-147. [ Links ]
SZEWZYK U, SZEWZYK R, MANZ W and SCHLEIFER KH (2000) Microbiological safety of drinking water. Annu. Rev. Microbiol. 54 81-127. [ Links ]
TAJIMA A, SAKURAI K and MINAMIYAMA M (2007) Behavior of pathogenic microbes in a treated wastewater reuse system and examination of new standards for the reuse of treated wastewater. Environ. Monit. Assess. 129 43-51. [ Links ]
VESPER S, McKINSTRY C, HARTMANN C, NEACE M, YODER S and VESPER A (2008) Quantifying fungal viability in air and water samples using quantitative PCR after treatment with propidium monoazide (PMA). J. Microbiol. Methods 72 180-184. [ Links ]
Received 26 August 2009; accepted in revised form 10 December 2009.
* To whom all correspondence should be addressed.
+574 219 6571; fax: +574 219 6571;
e-mail: rmagudelo123@hotmail.com














