Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
Southern African Journal of Critical Care (Online)
versión On-line ISSN 2078-676X
versión impresa ISSN 1562-8264
South. Afr. j. crit. care (Online) vol.37 no.3 Pretoria nov. 2021
http://dx.doi.org/10.7196/SAJCC.2021.v37i3.495
SHORT REPORT
A retrospective evaluation of a multiplex polymerase chain reaction test directly applied to blood for the management of sepsis in the critically ill
S OmarI; S MurphyII; R GheevargheseIII; N PoppletonIV
IMB ChB, FC Path (Chem) SA, DA (SA), Critical Care SA; Critical Care, School of Clinical Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
IIMB BCh, DCH (SA), FCPaed (SA), Cert Crit Care (SA) MScMed (Bioethics & Health Law); Critical Care, School of Clinical Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
IIIMB ChB, DCH (SA), FCPaed (SA), Cert Crit Care (SA); Critical Care, School of Clinical Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
IVMB ChB Critical Care, School of Clinical Medicine, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
ABSTRACT
BACKGROUND. Blood culture (BC) is the established gold standard for microbiological diagnosis of bloodstream infection (BSI); however, its sensitivity is poor.
OBJECTIVES. The primary objective was to determine the sensitivity and specificity of the Magicplex Sepsis Real-time Test, a multiplex polymerase chain reaction test (mPCR), and BC to detect BSIs. Secondary outcomes included determining the prevalence of BSIs.
METHODS. A retrospective review of a technical evaluation of the mPCR. Patients requiring BC had a blood sample collected for mPCR.
RESULTS. The respective sensitivity and specificity of mPCR for the detection of BSI were 50% (n=7/14) and 58% (n=18/31), while the sensitivity
and specificity using BC were 36% (n=5/14) and 68% (n=21/31), respectively. The addition of mPCR to BC increased BSI detection during sepsis from 36% to 64%.
CONCLUSION. The use of mPCR directly applied to blood may increase the detection of micro-organisms associated with BSIs in critically ill patients requiring BC investigation.
Keywords: polymerase chain reaction; PCR; sepsis; bloodstream infections; bood culture; critically illness; ICU.
Sepsis is defined as life-threatening organ dysfunction due to a dysregulated host response to infection.[1] The clinical appreciation of organ dysfunction is straightforward; however, pathogen identification is challenging. Early, appropriate antibiotic use is assessed as Grade 1b evidence.[2] Blood culture (BC) may have relatively poor sensitivity, as low as 32%.[3] The time to pathogen identification using BC is 48 - 72 hours, while the time to a negative BC is longer, 3 - 5 days.[4] BC contamination adds to clinical interpretation complexity.[5]
Amplification during polymerase chain reaction (PCR) allows detection of minuscule amounts of pathogen DNA within 4 - 6 hours.[6] The Magicplex Sepsis Real-time Test (Seegene, South Korea), a multiplex polymerase chain reaction test (mPCR), requires 1 mL of whole blood and has a 3 - 6-hour turnaround time. It identifies 90 pathogens, 27 at a species level and 3 resistance genes (mecA, vanA, vanB).[7]A technical evaluation of mPCR was performed in a multidisciplinary tertiary level intensive care unit (ICU).
Methods
Study design
A retrospective review of the technical evaluation and ICU database was performed. The mPCR's technical evaluation took place from 1 June to 31 July 2019. Approval was obtained from the Human Research Ethics Committee (HREC) of the University of the Witwatersrand (ref. no. M200251). Owing to the retrospective nature of the study, the requirement for informed consent was waived.
Study procedure
During the evaluation period, patients in whom infection was suspected, and from whom BCs were collected, were included. An additional blood sample was collected for the mPCR during the aseptic procedure. The ICU patient database was then cross-searched for clinical information, laboratory information, and antimicrobial prescriptions.
Blood cultures
BCs in the ICU are collected under strict aseptic technique as standard procedure (20 mL blood).
mPCR blood assay
All patients having BCs collected had an additional 4 mL ethylenediaminetetraacetic acid (EDTA) tube filled with blood during the procedure. Blood samples were directly processed according to the manufacturer's instructions to yield microbial nucleic acid material.
Microorganism identification followed three steps. The first PCR step identified Gram-positive bacteria and three resistant genes in one tube, and Gram-negative bacteria and six fungi in a second tube. The second PCR step identified organisms at a genus level: Streptococcus, Staphylococcus, and Enterococcus spp. in the first tube; Gram-negative bacteria group A (GNB-A) and GNB-B in the second tube; and fungi, vanA, vanB, and mecA resistance genes in the third tube. A third and final PCR was not performed.
GNB-A organisms included: Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia, Serratia marcescens, Bacteroides fragilis, and Salmonella typhi. GNB-B organisms included: Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis, Escherichia coli, Enterobacter cloacae, and Enterobacter aerogenes. Fungi included: Candida albicans, C. parapsilosis, C. glabrata, C. tropicalis, C. krusei and Aspergillus fumigatus.
Study definitions and research assumptions
Sepsis was defined using Sepsis-3 definition.[1] Infection was defined by a combination of clinical presence plus an associated biomarker increase above the upper reference limit (two of three markers: white cell count (WCC), C-reactive protein (CRP) and procalcitonin (PCT). BC and mPCR were considered concordant if the organism cultured from blood was an organism present in the corresponding mPCR group in the second PCR step. Appropriate clinical response was defined as a 40% reduction in either PCT or CRP over the following 48 hours. This was based on a daily decrease of 20% for 4 days reaching a 20%-of-peak threshold, allowing termination of antibiotics.[8] Appropriate antibiotic choices defined for the study included vancomycin for Enterococcus (provided the vanA and vanB genes were absent) and meropenem/imipenem for a GNB in group B with additional trimethoprim-sulfamethoxazole for group A. S. aureus-appropriate therapy included vancomycin or linezolid if the mecA gene was identified. Owing to limited fungal identification, only bacteria were analysed. Because of the retrospective nature of the study, mPCR results were not available to treating clinicians and antibiotic therapy was not actually changed; hence we refer to the comparative model as hypothetical.
Study outcomes
Primary outcome
To determine the sensitivity and specificity of mPCR and BC in detecting BSIs among patients with sepsis.
Secondary outcomes
To determine the prevalence of BSI, the concordance rates between BC and mPCR, the frequency of different implicated pathogens, and the frequency of inappropriate antibiotic therapy based on hypothetical knowledge from mPCR results availability at 12 hours compared with BC results at 72 hours.
Data analysis
Non-normal data were described using median and interquartile range (IQR). Independent medians and percentages were compared using the Mann-Whitney (7-test and χ2 test, respectively. Frequencies were described using numbers and percentages. Analysis was performed on Statistica version 13. 5.0.17 (TIBCO Software, USA).
Results
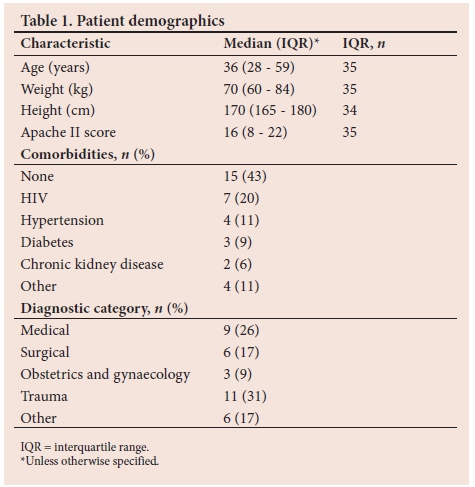
There were 45 episodes (35 patients) of clinically suspected infection or sepsis, each with simultaneous BC and mPCR testing performed. Fourteen patients met the definition of sepsis, 14 had a study-defined infection and 17 had clinically suspected infection. The patient demographics are provided in the Table 1. The number and percentage (n (%)) of suspected sites of infection were the lungs (n=19; 42.2%), abdomen (n=15; 33.4%), pelvis (n=5; 11.1%), skin/ soft tissue (n=2; 4.4 %) and other sites (n=4; 8.9% %).
Primary outcome
The sensitivity and specificity of mPCR for the detection of BSI among patients with sepsis was 50% (n=7/14) and 58% (n=18/31) respectively, while the sensitivity and specificity of BC was 36% (n=5/14) and 68% (n=21/31) respectively. When mPCR and BC positivity rates were compared, mPCR was more likely to detect a positive BSI, relative risk 1.82 (confidence interval 1.01 - 3.27). Three positive BCs among patients without sepsis were due to contamination and were included in the BC-negative group.
Secondary outcomes
Prevalence of bloodstream infections
The prevalence of BSI using BC in this study was 27%. A combination of BC and mPCR yielded the highest prevalence (Table 2). The concordance rates between BC and mPCR are provided in Table 2.
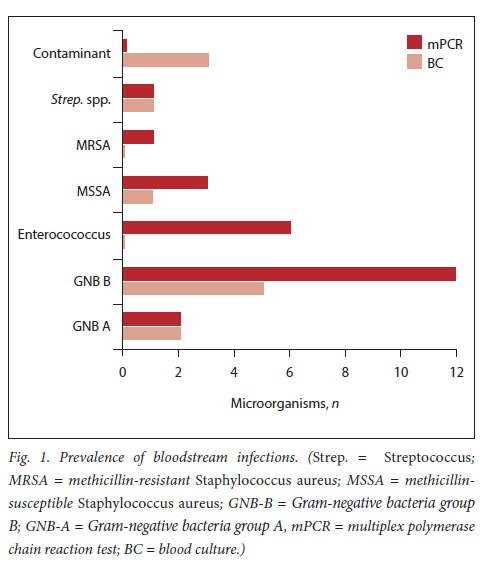
There were 14 microorganisms (2 fungal) cultured on blood and 25 identified on mPCR. Gram-negative bacteria were the most frequently identified bacterial pathogens (Fig. 1). Three contaminants were cultured on blood; two coagulase-negative Staphylococcus, and one Micrococcus spp.
Hypothetical comparative model
Availability of mPCR results was assumed at 12 hours post collection (mPCR 12h) compared with actual BC results at 72 hours (BC 72h). Two patients (8%) in the mPCR 12h group would have been on inappropriate antimicrobial therapy compared with 6 (23%) in the BC 72h group (p=0.25).
Discussion
In this technical evaluation, the sensitivity for detecting BSI in critically ill adult patients with sepsis in the ICU was 50% for mPCR and 36% for BC. A study by Carrara et al[9]showed that the same mPCR kit had a sensitivity of 65%. They included hospitalised patients and sepsis was not explicitly defined. A combination of mPCR and BC demonstrated a 68% prevalence of BSI compared with 64% in our study. Zboromyrska et al.[10] demonstrated a sensitivity of 65% with the mPCR test. However, their clinical assessment was not defined. The combination of mPCR and BC resulted in a 64% prevalence of BSI.[101 Ljungström et al.[11] evaluated the mPCR in emergency department suspected sepsis. They found a sensitivity of 64% using a rigorous clinical definition. Ziegler et al.[12] evaluated the mPCR kit's technical aspects without defined clinical outcomes. They found a 46% prevalence of BSI using BC and mPCR.
A systematic review illustrated the lack of a uniform reference against which the mPCR test (Magicplex) has been evaluated.[13] BC sensitivity and poorly defined clinical outcomes limit the interpretation of current data.
There was a high prevalence (27%) of BSI diagnosed by BC in our study. Previously published prevalences varied between 10 and 17%.[7,10-121 Our study was performed in a low- to middle-income country and may reflect a higher sepsis burden.[14]
Another advantage of mPCR was the low (6%) incidence of BC contaminants. This is in keeping with other published data showing a higher contamination rate for BC (10%) compared with mPCR (4.8%).[8]
The real value of mPCR may be when the pathogen yield is expected to be low, e.g. antimicrobial treatment.[15] Our study supports the complementary role of mPCR by nearly doubling the number of BC-diagnosed infections with only 1 mL of unincubated blood. We found a concordance rate between BC and mPCR of around 60%, similar to previous studies (60 - 80%).[9-11] It is this lack of complete concordance which facilitates the complementary role of mPCR to BC. In the hypothetical comparative model, mPCR in addition to BC could potentially reduce the number of patients on inappropriate therapy. This trend is hypothesis generating.
Study limitations and strengths
The retrospective, observational nature and small study size (report) are important limitations. We performed 2 of the 3 PCR steps, limiting a more direct comparison of microorganisms identified. However, we used established clinical definitions with objective measures to support clinically defined outcomes in a specific clinical setting (ICU).
Conclusions
The use of mPCR directly applied to blood may increase the detection of microorganisms associated with BSIs in critically ill patients requiring BC investigation.
Declaration. Results from this study were shared with staff members in the ICU in an informal presentation.
Acknowledgements. We would like to thank Donald van der Westhuizen, Business Manager for Molecular Diagnostics at Inqaba Biotec, for assistance with the technical training.
Author contributions. Authors contributed as follows to the conception or design of the work; the acquisition, analysis, or interpretation of data for the work; and drafting the work or revising it critically for important intellectual content: SO 45%, NP 35%, RG 5%, SM 15%. All authors approved the version to be published and agreed to be accountable for all aspects of the work.
Funding. No funding was received for this study owing to its retrospective nature. All time required by the investigators was volunteered. Printing and stationary costs were covered by the investigators in their individual capacity. Publication costs were covered by the investigators in their individual capacity.
Conflicts of interest. None.
References
1. Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315(8):801. https://doi.org/10.1001/jama.2016.0287 [ Links ]
2. Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock. Crit Care Med 2017;45(3):486-552. https://doi.org/10.1097/CCM.0000000000002255 [ Links ]
3. Warhurst G, Dunn G, Chadwick P, et al. Rapid detection of health-care-associated bloodstream infection in critical care using multi pathogen real-time polymerase chain reaction technology: A diagnostic accuracy study and systematic review. Health Techno Assess 2015;19(35):1-142. https://doi.org/10.3310/hta19350 [ Links ]
4. Opota O, Croxatto A, Prod'hom G, Greub G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin Microbiol Infect 2015;21(4):313-322. https://doi.org/10.1016/j.cmi.2015.01.003 [ Links ]
5. Weinstein MP. Blood culture contamination: Persisting problems and partial progress. J Clin Microbiol 2003;41(6):2275-2278. https://doi.org/10.1128/jcm.4L6.2275-2278.2003 [ Links ]
6. Dark PM, Dean P, Warhurst G. Bench-to-bedside review: The promise of rapid infection diagnosis during sepsis using polymerase chain reaction-based pathogen detection. Crit Care Lond Engl 2009;13(4):217. https://doi.org/10.1186%2Fcc7886 [ Links ]
7. Florio W, Morici P, Ghelardi E, Barnini S, Lupetti A. Recent advances in the microbiological diagnosis of bloodstream infections. Crit Rev Microbiol 2018;44(3):351-370. https://doi.org/10.1080/1040841x.2017.1407745 [ Links ]
8. Azzini AM, Dorizzi RM, Sette P, et al. A 2020 review on the role of procalcitonin in different clinical settings: An update conducted with the tools of the evidence based laboratory medicine. Ann Transl Med 2020;8(9):610-610. https://doi.org/10.21037%2Fatm-20-1855 [ Links ]
9. Carrara L, Navarro F, Turbau M, et al. Molecular diagnosis of bloodstream infections with a new dual-priming oligonucleotide-based multiplex PCR assay. J Med Microbiol 2013;62(Pt11):1673-1679. https://doi.org/10.1099/jmm.0.064758-0 [ Links ]
10. Zboromyrska Y, Cillóniz C, Cobos-Trigueros N, et al. Evaluation of the MagicplexTM Sepsis Real-time Test for the rapid diagnosis of bloodstream infections in adults. Front Cell Infect Microbiol 2019;9:1-8. https://doi.org/10.3389%2Ffcimb.2019.00056 [ Links ]
11. Ljungström L, Enroth H, Claesson BE, et al. Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis. BMC Infect Dis 2015;15(1):199. https://doi.org/10.1186/s12879-015-0938-4 [ Links ]
12. Ziegler I, Fagerström A, Strâlin K, Mölling P. Evaluation of a commercial multiplex PCR assay for detection of pathogen DNA in blood from patients with suspected sepsis. PLoS ONE 2016;11(12).https://doi.org/10.1371/journal.pone.0167883 [ Links ]
13. Chang S-S, Hsieh W-H, Liu T-S, et al Multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis: A systemic review and meta-analysis. PloS ONE 2013;8(5): https://doi.org/10.1371/journalpone.0062323 [ Links ]
14. Becker JU, Theodosis C, lacob ST, Wira CR, Groce NE. Surviving sepsis in low-income and middle-income countries: New directions for care and research. Lancet Infect Dis 2009;9(9):577-582. https://doi.org/10.1016/s1473-3099(09)70135-5 [ Links ]
15. Timsit J-F, Ruppé E, Barbier F, Tabah A, Bassetti M. Bloodstream infections in critically ill patients: An expert statement. Intensive Care Med 2020;46(2):266-284. https://doi.org/10.1007/s00134-020-05950-6 [ Links ]
 Correspondence:
Correspondence:
S Omar
shahedicu@gmail.com
Accepted 4 August 2021
Contribution of the study. Our data add to a growing body of evidence indicating that mPCR applied directly to blood prior to incubation increases the detection of pathogenic bacteria among hospitalised patients for whom blood cultures are performed for suspected infection. Our study was performed in a low-to-middle income country with a higher sepsis prevalence, a greater burden of multidrug-resistant organisms and clinically defined sepsis. This strengthens the robustness and generalisability of this body of evidence.














