Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
Southern African Journal of Critical Care (Online)
versión On-line ISSN 2078-676X
versión impresa ISSN 1562-8264
South. Afr. j. crit. care (Online) vol.31 no.1 Pretoria jun. 2015
http://dx.doi.org/10.7196/SAJCC185
ARTICLE
Organisms cultured and resistance patterns seen in a secondary referral centre ICU and burns unit
B GreatorexI; G OosthuizenII
IBMBS, FRCA; Departments of Intensive Care and Surgery, Edendale Hospital, Pietermaritzburg, KwaZulu-Natal, South Africa
IIMB ChB, FCS (SA); Departments of Intensive Care and Surgery, Edendale Hospital, Pietermaritzburg, KwaZulu-Natal, South Africa
ABSTRACT
BACKGROUND: Infections are common in intensive care units (ICUs) and burns units. Empiric antibiotic therapy is often required, and as such it is important to have a good knowledge of the resident organisms in these departments. Antibiotic resistance is becoming an increasing problem both internationally and in South Africa (SA) and it is important to monitor organism sensitivity.
OBJECTIVES: To establish the spectrum and sensitivity of nosocomial pathogens in an SA government referral hospital ICU and burns unit.
METHODS: We report the findings from a retrospective audit of all cultures sent from the ICU and burns unit of an SA urban hospital for a 6-month period between January and June 2008.
RESULTS: The results showed a prevalence of Gram-negative organisms in the ICU department, in particular Klebsiella pneumoniae and Escherichia coli. There was a prevalence of Gram-positive organisms in the burns unit. Overall resistance to co-amoxiclav and erythromycin was found to be high (49% and 53%, respectively), resistance to ciprofloxacin and gentamicin was moderate (30% and 35%, respectively) and resistance to piperacillin-tazobactam and the carbapenems remained low (21% for piperacillin-tazobactam, 2% for ertapenem and 19% for meropenem). When looking at individual species, it was noted that K. pneumoniae had high resistance to ampicillin (97%), moderate resistance to co-amoxiclav and ciprofloxacin (35% and 43%, respectively) and low resistance to piperacillin-tazobactam, ertapenem, meropenem and colistin (12%, 0%, 5% and 0%, respectively). E. coli was seen to have high resistance to ampicillin (79%), but low resistance to co-amoxiclav (4%), ciprofloxacin (9%), piperacillin-tazobactam (0%), ertapenem (0%), meropenem (4%) and colistin (0%).
CONCLUSION: This study demonstrates the prevalence of Gram-negative organisms in an SA government hospital ICU. It also demonstrates the presence of resistance mechanisms in the organisms cultured for almost all available classes of antibiotics, albeit some at low levels. The development of multi- and pan-resistant pathogenic organisms is both an SA and worldwide problem. In particular, the threat posed by resistant Gram-negative bacteria is likely to manifest itself in ICUs where septic patients unresponsive to standard antimicrobial regimens will inevitably end up. Frequent assessment of resistance patterns and appropriately designed empirical treatment protocols must remain a priority for all critical care departments.
Invasive infections are common in intensive care units (ICUs) and burns units, and mortality from severe sepsis in the critical care setting ranges from 28 to 50%.[1] Antibiotics are commonly prescribed to critically ill patients. The use of antibiotics that are ineffective against the causative organism is associated with increased morbidity and mortality, and is an unnecessary additional strain on what are often stretched healthcare resources.[2,3] A study from 2005 looking at prescribing practices in ICUs across South Africa (SA) identified inappropriate empirical antibiotic choice in 54.9% of patients.[3] Inappropriate use can also result in the development of antibiotic resistance among bacteraemic pathogens.[4] As such, it is important to ensure that empirical antibiotic choice is based on good knowledge of the resident organisms in critical care departments. It is also well known that resistance among pathogens is increasing rapidly both internationally and in SA.[5,6] New forms of antibiotic resistance can spread between countries with ease. Of particular concern are the so-called 'ESKAPE' pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.[7] Of the aforementioned pathogens, while there is still concern around methicillin-resistant Staphylococcus aureus (MRSA), it is infections secondary to the Gram-negative bacteria that are causing greater concern due to the existence of pan-resistant strains and the paucity of novel antibiotics.[8] A recently published audit from an ICU in Durban, South Africa, revealed that from samples sent from ventilated patients, 62% of the positive results were Gram-negative bacteria. Extended spectrum β-lactamase production was found in 19% of Escherichia coli cultured and 25% of Klebsiella spp. isolates.[9] Over time, as antibiotics are used within a department, the resident organisms will adapt and develop resistance mechanisms such as the production of β-lactamases. Therefore, it is important to monitor organisms and their spectrum of resistance on a regular basis.
Objectives
To establish the spectrum and sensitivity of nosocomial pathogens in an SA government referral hospital ICU and burns unit.
Methods
We report the findings from a retrospective audit of all cultures sent from the ICU and burns unit of Edendale Hospital, Pietermaritzburg, SA, for a 6-month period between January and June 2008. All culture and sensitivity testing was carried out at an on-site laboratory. The data that were assessed were kept in hard-copy form in the microbiology department of the hospital.
Edendale is a government hospital with 900 beds, which included at the time of the audit a 6-bed adult ICU and an 8-bed acute burns unit. There were no isolation rooms. There were no ventilators in the burns unit; patients requiring invasive respiratory support were admitted to the ICU. It was not policy in either the ICU or burns unit to send surveillance cultures at the time the audit was performed. As such, all specimens were sent in response to suspected infection. Specifically, neither tracheal aspirates nor wound swabs were sent routinely. Bronchoscopy and bronchoalveolar lavage were not possible for sputum sample collection in the ICU.
All data were recorded in an Excel (Microsoft, USA) spreadsheet, including those samples with no growth. The data that were recorded included the type of sample sent, any organisms grown and their susceptibility to any antibiotics tested. Any results which were from the same patient, during the same visit and which grew the same organism were only counted once. Results for which the genus was given, but not the specific name, were categorised separately under the title spp. (for example, Klebsiella spp.).
Results
Table 1 shows the organisms cultured from patients in the ICU. The total number of positive cultures was 127. Gram-negative organisms predominated, in particular K. pneumoniae, which represented 27% of cultured isolates, E. coli (22%) and A. baumannii (17%). Table 2 shows the organisms cultured from patients in the burns unit. The total number of positive cultures was 20. There was prevalence for Gram-positive organisms, in particular S. aureus and S. epidermidis.


Table 3 shows the number and type of cultures sent from the ICU during the 6-month period, and the percentage of cultures that came back positive. Blood was the most common sample type sent (158 samples), of which 60 were growth positive. Despite fewer samples of sputum and tracheal aspirate being sent (76 samples), the number of samples that were growth positive was higher than for blood, at 68. The high percentage of positive growth seen in sputum and tracheal aspirates may reflect the techniques used to collect the samples. The number of growth-positive wound swabs was also high, at 62, a proportion of which almost certainly represented skin commensals. Table 4 shows the number and type of cultures sent from the burns unit. As might be expected, the majority of samples sent were wound swabs (38 samples). Similarly to what was seen in the ICU, a high proportion (84%) of wound swab cultures came back positive.
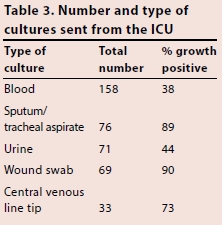
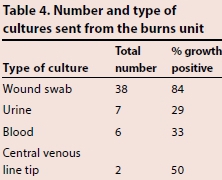
Table 5 demonstrates the resistance rates of each genus to all of the antibiotics routinely tested. Low levels of resistance were seen to linezolid (0%), amikacin (8%), vancomycin (10%) and colistin (12%). Moderate levels of resistance were seen to piperacillin-tazobactam (21%), ciprofloxacin (30%), gentamicin (35%) co-amoxiclav (49%) and the 3rd-generation cephalosporins cefotaxime and ceftazidime (49% and 44%, respectively). The level of resistance to the carbapenems varied from 2% for ertapenem up to 23% for imipenem.
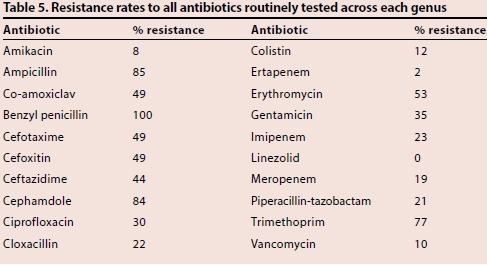
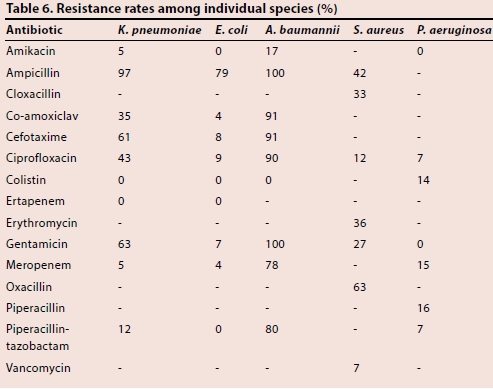
Table 6 demonstrates resistance rates among individual species. A. baumannii displayed the highest levels of resistance with 80% and 78% resistance to piperacillin-tazobactam and meropenem, respectively. By comparison, 12% of K. pneumoniae was resistant to piperacillin-tazobactam, while 5% was resistant to meropenem.
Discussion
Nosocomial infections cause significant morbidity and mortality among patients. Of these infections, those affecting the lower respiratory tract, urinary tract, bloodstream and postsurgical account for the majority. [10] The increasing resistance rate seen in nosocomial pathogens combined with a paucity of new antimicrobials presents a serious challenge for physicians.[11] There are concerns that we are entering a post-antibiotic era where the treatment of previously curable infections becomes impossible. It is likely that this phenomenon will be seen most acutely in ICUs, as they are often the final destination of patients with septic shock who are not responding to initial, empirical antimicrobial therapy.
The organisms that were grown from the samples sent during the 6 months of this audit represent a combination of community-acquired and nosocomial pathogens. Distinguishing which are hospital acquired is not possible from our data. However, knowing the patterns of resistance across all pathogenic species helps us to establish more effective empirical antibiotic protocols.
Organisms grown
In this study, Gram-negative bacteria made up the majority of the growth-positive cultures (114 cultured organisms of a total of 127) in the ICU, the three most prevalent of which were K. pneumoniae (26.8% of cultured isolates), E. coli (22.0%) and A. baumannii (17.3%). S. aureus was the only Gram-positive organism cultured in the ICU in significant numbers (10.2%). The preponderance for Gram-negative organisms is consistent with the findings of Brink et al.[4] for private institutions in SA in 2008. They found that K. pneumoniae made up 24.9% of cultured isolates, while E. coli and A. baumannii made up 18.5% and 7.4%, respectively. S. aureus made up 24.6%. Notably, unlike Brink et al.,[4] we saw very few Enterobacter spp. (n=7 positive cultures, all E. cloacae). We also saw very few Enterococci spp. (n=8), none of which were resistant to vancomycin. In 2013, when looking at an SA trauma ICU, Ramsamy et al.[9]found that Gram-negative organisms made up the majority of the cultured organisms. However, in their study, Gram-positive organisms made up a much bigger proportion of the total (119 out of 323). This emphasises the importance of knowing the common pathogenic organisms in a unit when designing empirical antimicrobial protocols.
In contrast to the ICU, Gram-positive organisms were the most common organism cultured in the burns unit, in particular S. aureus. This isn't surprising perhaps, given the fact that most of the cultures were wound swabs and therefore possibly represented skin flora rather than invasive pathogens. However, immediate colonisation of burns by patients' normal skin flora is well documented and, depending on the individual organism's virulence, can often lead to an invasive burn wound infection.[12]
Specific organisms of concern
K. pneumoniae (n=34)
K. pneumoniae showed similarly high rates of resistance to ampicillin, co-amoxiclav, ciprofloxacin and the 3rd-generation cephalosporins to those seen by Brink et al.[4]in private institutions. In contrast to their findings, however, we found low levels of resistance to piperacillin-tazobactam (12% compared with 40%). Resistance to the carbapenems was also comparably low. Despite the increasing prevalence of carbapenemase-producing Klebsiella isolates in Europe (8.3% in 2013)[13] and the US (11% for all Klebsiella spp. in 2013),[14] documented rates of carbapenem resistance in Africa remain low, at between 0 and 4% (although there is a paucity of data).[15]
E. coli (n=28)
Although resistance to ampicillin among E.coli isolates was also high (79%), they were surprisingly sensitive to all other classes of antimicrobials (4%, 9% and 8% for co-amoxiclav, ciprofloxacin and cefotaxime, respectively). This is in contrast to the findings of both Brink et al.,[4]who documented resistances of 37%, 20% and 10%, respectively, for the same three antimicrobials, and the World Health Organization (WHO)[15] data for 2014, which showed resistance in Africa to fluoroquinolones of between 34 and 53% in invasive isolates and resistance to 3rd-generation cephalosporins of between 28 and 36%. There was no documented resistance to either piperacillin-tazobactam or ertapenem, and only 4% to meropenem.
A. baumannii (n=22)
Of all the organisms cultured from patients in the Edendale ICU, A. baumannii exhibited the most extensive range of antimicrobial resistance. This is in contrast to Brink et al.,[4]who saw significantly lower levels of resistance to ciprofloxacin (90% compared with 36%), 3rd-generation cephalosporins (91% compared with 43%) and piperacillin-tazobactam (80% compared with 42%). Even more concerning, however, was the level of resistance to meropenem (78%). The Centers for Disease Control reported in 2013 that 63% of all nosocomial A. baumannii infections in the USA were multidrug resistant. The only effective antimicrobial tested at Edendale was the polymyxin, colistin (0% resistance). The use of colistin as monotherapy, part of combination therapy and via novel routes such as nebulisation has seen a renewed interest in recent years. While there have been doubts in the past about the virulence of A. baumannii, given the ability of micro-organisms to transfer resistance genes between genus, the presence of bacteria with such an extensive spectrum of resistance is a large concern.
S. aureus (n=20)
The overall resistance to cloxacillin or oxacillin, thus conferring methicillin-resistant status, was 46%. This is within the range reported by the WHO[15] for the African continent in 2014 (12 -80%). However, it is slightly greater than that seen by Brink et al.[4] (36%) and significantly higher than the EU average for 2013 of 18% (although the EU intercountry variation is 1 - 60%). It should be noted that MRSA rates in both the US and Europe have been falling over the past 6 years, possibly secondary to focused national infection control campaigns.[13,14] In line with most other studies, resistance to vancomycin was low at 7%.
P. aeruginosa (n=12)
Resistance to ciprofloxacin, gentamicin and piperacillin-tazobactam was low at 7%, 0% and 7%, respectively. This is in marked contrast to Brink et al.,[4] who saw resistance in the private sector of 46% for ciprofloxacin, and 48% for both amikacin and piperacillin-tazobactam. Resistance to both colistin and meropenem was around 15%, again lower than Brink et al. The CDC have reported that ~8% of all healthcare-associated infections were caused by P. aeruginosa and that 13% of these were multi-drug resistant.
Study limitations
These data are historical, and while useful in guiding empirical antimicrobial protocols at the time they were collected, it is likely that the spectrum of antimicrobial resistance has changed significantly since. To guide current protocols, further data should be collected. Data regarding length of stay before cultures were taken was not collected, so no distinction can be made between community- and hospital-acquired infections. Distinguishing the hypermetabolic response to burns from sepsis is challenging, and as such, the wound swabs sent from the burns unit may often reflect colonising bacteria rather than invasive pathogens.
Conclusions
This study demonstrates the prevalence of Gram-negative organisms in an SA government hospital ICU. It also demonstrates that while at low levels for certain classes of antibiotics, specifically carbapenems, resistance mechanisms exist in the organisms cultured for almost all available classes of antibiotics. The development of multi- and pan-resistant pathogenic organisms is both an SA and worldwide problem. In particular, the threat posed by resistant Gram-negative bacteria is likely to manifest in ICUs where septic patients unresponsive to standard antimicrobial regimens will inevitably end up. Frequent assessment of resistance patterns and appropriately designed empirical treatment protocols must remain a priority for all critical care departments.
References
1. Sablotzki A, Muhling J, Czeslick E. Sepsis and multiple organ failure: Update of current therapeutic concepts. Anaesthesiol Intensivmed Notfallmed Shmerzther. 2005;40(9):511-520. [http://dx.doi.org/10.1055/s-2005-870104] [ Links ]
2. Colardyn F. Appropriate and timely empirical antimicrobial treatment of ICU infections: A role for carbapenems. Acta Clin Belg 2005;60(2):51-62. [http://dx.doi.org/10.1179/acb.2005.011] [ Links ]
3. Paruk F, Richards G, Scribante J, Bhagwanjee S, Mer M, Perrie H. Antibiotic prescription practices and their relationship to outcome in South African intensive care units: Findings of the Prevalence of Infection in South African Intensive Care Units (PISA) Study. S Afr Med J 2012;102(7):613-616. [ Links ]
4. Brink A, Moolman J, Da Silva MC, Botha M, National Antibiotic Surveillance Forum. Antimicrobial susceptibility profile of selected bacteraemic pathogens from private institutions in South Africa. S Afr Med J 2007;97(4):273-279. [ Links ]
5. Brink A, Feldman C, Richards G, Moolman J, Senekal M. Emergence of extensive drug resistance (XDR) among gram negative bacilli in South Africa looms nearer. S Afr Med J 2008;98(8):586-592. [ Links ]
6. Jones RN. Resistance patterns among nosocomial pathogens: Trends over the past few years. Chest 2001;119(2 Suppl):S397-S404. [ Links ]
7. Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J Infect Dis 2008;197:1079-1081. [http://dx.doi.org/10.1086/533452] [ Links ]
8. Pop-Vicas A, Opal SM.The impact of multi-drug resistant gram-negative bacilli in the management of septic shock. Virulence 2014;5(1):206-212. [http://dx.doi.org/10.4161/viru.26210] [ Links ]
9. Ramsamy Y, Muckart DJ, Han KS. Microbiological surveillance and antimicrobial stewardship minimise the need for ultrabroad-spectrum combination therapy for treatment of nosocomial infections in a trauma intensive care unit: An audit of an evidence-based empiric antimicrobial policy. S Afr Med J 2013;103(6):371-376. [http://dx.doi.org/10.7196/samj.6459] [ Links ]
10. Dusé AG. Infection control in developing countries with particular emphasis on South Africa. South Afr J Epidemiol Infect 2005;20(2):37-41. [ Links ]
11. Pertrosillo N, Capone A, Di Bella S, Taglietti F. Management of antibiotic resistance in the intensive care unit setting. Expert Rev Anti Infect Ther 2010;8(3):289-302. [http://dx.doi.org/10.1586/eri.10.7] [ Links ]
12. Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev 2006;19(2):403-434. [http://dx.doi.org/10.1128/CMR.19.2.403-434.2006] [ Links ]
13. European Centre for Disease Prevention and Control. Summary of the latest data on antibiotic resistance in the European Union. 2014. http://ecdc.europa.eu/en/eaad/Pages/antibiotics-data-reports.aspx (accessed 1 February 2015). [ Links ]
14. US Department of Health and Human Services Centre for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed 1 February 2015). [ Links ]
15. World Health Organization. Antimicrobial Resistance Global Report on Surveillance. 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed 1 February 2015). [ Links ]
 Correspondence:
Correspondence:
B Greatorex
benjamingreatorex@hotmail.com














