Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
Journal of the South African Veterinary Association
versión On-line ISSN 2224-9435
versión impresa ISSN 1019-9128
J. S. Afr. Vet. Assoc. vol.95 no.1 Pretoria 2024
http://dx.doi.org/10.36303/JSAVA.597
ORIGINAL RESEARCH
A novel loop-mediated isothermal amplification (LAMP) assay to diagnose feline panleukopenia
RA BakdeI; RL RathishI; L JohnII; PM DeepaI; K VijayakumarIII
IDepartment of Veterinary Epidemiology and Preventive Medicine, College of Veterinary and Animal Sciences, Kerala Veterinary and Animal Sciences University, India
IIDepartment of Veterinary Biochemistry, College of Veterinary and Animal Sciences, Kerala Veterinary and Animal Sciences University, India
IIICollege of Veterinary and Animal Sciences, Kerala Veterinary and Animal Sciences University, India
ABSTRACT
Protoparvovirus carnivoran1, known commonly as feline panleukopenia virus (FPV) is a highly contagious and environmentally stable parvovirus of domestic as well as wild felids. A rapid and robust diagnostic tool will aid in implementing prompt treatment and control measures.
A loop-mediated isothermal amplification (LAMP) as a point-of-care diagnostic tool for diagnosing feline panleukopenia was standardised using faecal samples of cats. The assay will reduce the cost and time required to diagnose feline panleukopenia. A set of two outer primers (F3 and B3) and two inner primers (FIP and BIP) were designed to target the viral polypeptide (VP2) gene of FPV. Optimisation of the LAMP reaction was done at 60 °C for one hour after an initial denaturation at 95 °C for five minutes. Visualisation of the result based on the addition of SYBR Green 1 dye offered an easy and reliable diagnosis.
The detection limit of the standardised LAMP assay was as low as 1.25 ng/|J of the target DNA. Species specificity of the LAMP primers revealed no amplification of the non-target DNA of any other species except that of the canine parvovirus DNA template. DNA extracted from 100 PCR-positive and 20 PCR-negative faecal samples were subjected to the standardised assay and compared with PCR.
Analysis of the results revealed that the LAMP assay was 100% sensitive and 90% specific compared to PCR. The LAMP assay could be a reliable tool for the point-of-care diagnosis of feline panleukopenia in limited resource settings.
Keywords: feline panleukopenia, LAMP, point-of-care diagnosis, domestic and wild felids
Introduction
Feline panleukopenia is a viral disease affecting domestic and wild felids caused by a ubiquitous single-stranded parvovirus of the genus Protoparvovirus and it includes canine parvovirus, mink enteritis virus and blue fox parvovirus (Rehme et al. 2022). The virus remains persistent in environmental conditions for over a year and can be transmitted through fomites or asymptomatically infected cats (Bergmann et al. 2019). A high mortality rate has been reported in kittens younger than five months old following an acute phase of illness (Yang et al. 2022). Thus, prompt diagnosis in clinical settings may play a pivotal role in reducing the mortality associated with feline panleukopenia virus (FPV). Though cat-rearing has been in vogue since ancient times in Kerala, India, adopting pure-bred cats as pets is relatively new (Vincy & Tresamol 2023). Vaccination of cats is often neglected and has given rise to an increased occurrence of infectious diseases like feline panleukopenia and feline rhinotracheitis in Kerala (Datta et al. 2021). Conventionally, diagnosis of feline panleukopenia involves polymerase chain reaction (PCR), which requires expensive instrumentation. Enzyme-linked immunosorbent assay (ELISA) is a lengthy process, whereas isolating viruses may also take a considerable amount of time and effort. Also, the necessity for expensive visualisation methods reduces their utility in clinical facilities. The SNAP test is a newly developed diagnostic test based on antigen detection (Jacobson et al. 2021). The test is rapid and reliable but not as sensitive as DNA amplification methods. Therefore, the development of rapid diagnostic methods that can be adapted in clinical facilities with limited resources is needed.
LAMP is a nucleic acid detection method that can be completed in under an hour in isothermal conditions (Notomi et al. 2000). The LAMP assay was based on two inner primers and two outer primers that recognised six distinct regions on the target gene and depends on the strand displacement activity of Bacillus stearothermophillus (Bst) polymerase resulting in the formation of highly stable loop structures (Kang et al. 2006).
LAMP offers several benefits over traditional nucleic acid detection methods in not requiring expensive laboratory set-up and gel documentation systems. The results of the LAMP reaction can be confirmed by observing a ladder-like pattern on agarose gel electrophoresis. Results can also be observed by assessing the turbidity due to the release of magnesium pyrophosphate (Parida et al. 2008) or by using intercalating dyes such as SYBR Green 1 (Lai et al. 2021).
The objective of the present study was to standardise LAMP for the detection of FPV from the faecal samples of cats and to ascertain the sensitivity and specificity of LAMP in comparison with traditional PCR.
Materials and methods
Sample collection and template DNA
A total of 120 cats showing signs suggestive of feline panleukopenia presented to the Teaching Veterinary Clinical Complex (TVCC), Pookode Kerala, India formed the subject of the study. Suspected cases were identified based on the history and clinical signs. Microscopy of faecal samples was done to rule out intestinal parasites, and haematology was employed to detect panleukopenia. Faecal samples were collected for DNA purification using sterile swabs and stored at -20 °C. DNA was purified using DNAzol™ reagent (Chomczynski et al. 1997). The DNA isolated from a commercially available trivalent live vaccine containing FPV was used as a positive control template for PCR.
PCR and sequencing of amplicons
The PCR reaction was carried out on the DNA extracted from faecal samples using primers FMF and FMR targeting the VP2 gene of the FPV (Mochizuki et al. 1996). The amplicon from the positive control and another from the clinical sample were sequenced using automated Sanger dideoxy nucleotide sequencing by outsourcing to M/s AgriGenome Labs Pvt. Ltd., Kochi. Sequences on subjecting to global sequence comparisons using the basic logical alignment search tool (BLAST) hosted by the National Centre for Biotechnology Information (NCBI) confirmed the identity of the FPV in the isolate. The sequence was deposited at NCBI GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with Accession No. https://www.ncbi.nlm.nih.gov/nuccore/°%20MN549355.1/. The template DNA of the confirmed isolate was stored at -20 °C and used as the positive control for further testing of the LAMP assay.
Standardisation of LAMP
The LAMP reaction was standardised using a set of two inner (FIP, BIP) and two outer primers (F3, B3) designed using Primer ExplorerTM (Version 5) (http://primerexplorer.jp/lampv5e/index.html), targeting the VP2 gene of FPV. Details of the primer sequence are given in Table I.
Reagents for the LAMP assay were procured from New England Biologicals. Template DNA derived from the positive control was employed to optimise the LAMP reaction. In a 25 μl reaction volume, the mixture consisted of 1 μl of template DNA, 1 μl each of the forward inner primer (FIP) and backward inner primer (BIP) at 40 pmol each, and 1 ul each of the forward outer primer (F3) and backward outer primer (B3) at five pmol each. Additionally, the reaction included 2.5 μl of a 10X ThermoPol reaction buffer and 3.5 ul of 1.4 mmol/l deoxynucleotide triphosphates. The magnesium sulfate concentration was adjusted to a final level of 16 mM. All the ingredients were thoroughly mixed and heated at 95 °C for five minutes for initial denaturation. After this, 1 μl of 8 U Bst DNA polymerase (Cat No. M0275, New England Biolabs, USA) was added.
Experiments were conducted at four different temperatures: 55 °C, 60 °C, and 65 °C, with each reaction terminated at 80 °C for two minutes to identify the optimal reaction temperature. After determining the optimal reaction temperature, various reaction times, including 15, 30, 45, and 60 minutes, were tested.
In addition to temperature and reaction time optimisation, MgSO4 concentrations at 4, 8, and 16 mM, ThermoPol at 2 μl and 2.5 μl, and betaine at 0.8 M, 1 M, and 1.6 M were also tested.
Amplification could be detected from the turbidity resulting from the precipitation of magnesium pyrophosphate. To enhance visualisation, 1 μl of a 1:10 diluted 10,000x SYBR Green 1 dye was added at the end of the reaction (Hamburger et al. 2013). Successful amplification was confirmed by a colour change in the SYBR Green solution, transitioning from orange to apple green. Successful amplification revealed a ladder-like pattern on agarose gel electrophoresis on a 2% agarose gel. Final reaction mix is given in Table II.
Analytical sensitivity and specificity of LAMP assay
The sensitivity of LAMP primers was assessed by a five-fold serial dilution of the quantified positive control DNA for LAMP assay from 20 ng to 0.3125 ng. The DNA was quantified using Nanodrop 2000 Spectrophotometer™ (Thermofischer, USA).
To determine the species specificity of the LAMP primers, the assay was carried out using non-target DNA samples, which included whole DNA extracted from Cat (Felis cattus) blood, Escherichia coli, canine parvovirus, nasal swab positive for feline rhinotracheitis and scab from a case of swine pox. All the non-target DNAs were sourced from clinical samples submitted to our laboratory. The positive control and a non-template control were also run in parallel.
Comparison of LAMP and PCR
Diagnostic sensitivity and specificity of a test is the ability of the test to detect true positive and true negative cases in a given population. In the present study, assuming PCR as the gold standard diagnostic test for feline panleukopenia, the LAMP assay was evaluated. PCR and LAMP assays were done in all 120 suspected feline panleukopenia cases. The results were interpreted in a 2x2 contingency table and the diagnostic sensitivity, specificity, positive predictive value, and negative predictive value of the LAMP assay were calculated as per Thrusfield et al. (2018).
Ethical considerations
The animal study was reviewed and approved by the faculty research committee vide Order No. KVASU/DAR/Acad/ A3/35766/2018 of Kerala Veterinary and Animal Sciences University. Experiments were done on clinical specimens submitted to our laboratory by registered veterinarians. Written informed consent for participation was not obtained from the owners because the samples were collected from cats brought to the veterinary clinics managed by registered veterinary practitioners. The clinical samples were collected by the veterinarians after getting oral consent from the pet owners for the detection of pathogenic organisms in their pets.
Results
PCR and sequencing of amplicons
PCR and agarose gel electrophoresis showed that out of 120 faecal samples, 100 samples (83.33%) had detected FPV DNA while the remaining 20 were PCR-negative. The PCR amplification produced an approximately 700 bp amplicon as shown in Figure 1. It was sequenced and revealed a size of 698 bp. The BLAST analysis of the sequence showed that it was a sequence of FPV and clustered with FPV VP2 sequences reported from other parts of the world with 100% identity and zero "E" value. The sequence data was submitted to the National Centre for Biotechnology Information database with accession number MN549355.
Validation of LAMP primers
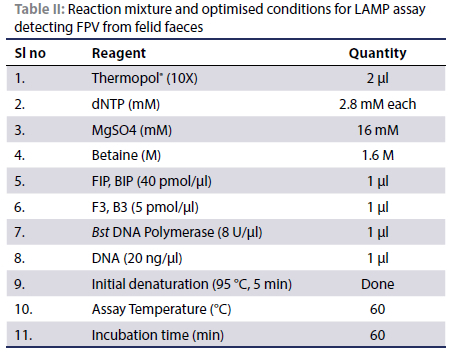
PCR was conducted on the positive control DNA using the forward and reverse outer primers (F3 and B3) designed for the LAMP assay. After agar gel electrophoresis, 200 bp long amplicons were obtained at the expected positions as shown in Figure 2. The successful amplification indicated that the primers were capable of amplifying the VP2 region under consideration.
Optimisation of the assay
The LAMP-based amplification of the VP2 gene of FPV was optimised using a commercial multivalent vaccine containing FPV as a positive control. Initial denaturation at 95 °C for five minutes was required for amplification. Amplification could be observed following incubation at 60, 63 and 65 °C for 60 minutes. No amplification was observed following incubation at 55 °C. Similarly amplification was unsuccessful at 15, 30 and 45 minutes. Ladder-like pattern was best viewed in agar gel electrophoresis when the reaction was carried out at 60 °C. The assay was optimised at 60 °C for 60 minutes.
Optimal visualisation was achieved at a magnesium sulfate concentration of 16 mM, 1.6 M betaine, and the addition of 2 μL of 10X thermopol buffer.
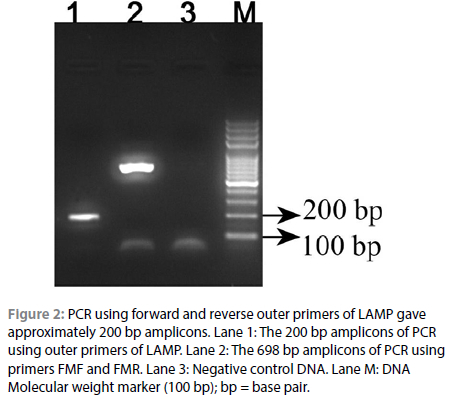
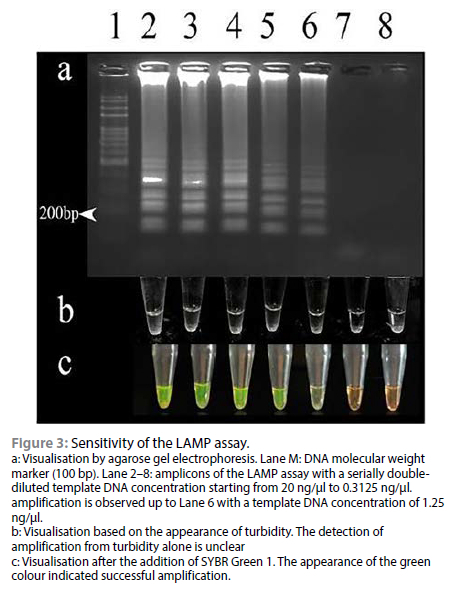
Turbidity was noted in all the positive vials upon completion. No turbidity was observed in non-template control as well as negative control. Positive LAMP reactions were also visualised after the addition of 1 μl of 1/10 diluted SYBR Green 1 to them. The solution turned green in the presence of amplicons and remained orange when there was no amplification, as in Figure 3c. A typical ladder-like pattern ending at about 200 bp was observed for the positive reactions as in Figure 3a Unambiguous differentiation of positive and negative reactions by observing turbidity was difficult, but was easy on using SYBR Green I.
Sensitivity and specificity of LAMP assay
The sensitivity of the LAMP assay standardised in this study was determined by amplifying a serial double dilutions of the positive control DNA starting from 20 ng up to a final concentration of 0.3125 ng. The positive reaction could be visualised up to a concentration of 1.25 ng/ul, as depicted in Figure 3.
The specificity of the FPV-LAMP assay was determined using the DNA of the domestic cat, Escherichia coli, feline rhinotracheitis, swine pox and canine parvovirus DNA as templates. Amplification was observed with the positive control and canine parvovirus DNA template indicated by the development of a green colour upon the addition of SYBR Green 1 dye. No amplification was observed with any other template DNA.
Comparison of LAMP and PCR
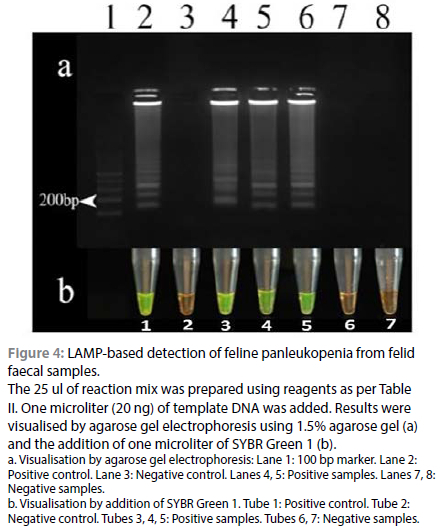
Out of the 120 DNA samples tested, all the 100 PCR-positive samples tested positive in LAMP, while two of the PCR-negative samples tested positive in LAMP. The remaining 18 samples were negative in both PCR and LAMP. The result of the LAMP assay is shown in Figure 4. The results of PCR and LAMP assay are summarised as a 2x2 contingency table in Table III. The positivity rate of the LAMP assay was 85%. In comparison with PCR, the diagnostic sensitivity and specificity of LAMP were found to be 100% and 90%, respectively. The positive and negative predictive values were found to be 98.04% and 100%, respectively. The accuracy of the test was 98.33%, indicating a good agreement between the tests.
Discussion
FPV is a highly contagious and environmentally stable parvovirus of domestic and wild felids. A rapid and robust diagnostic tool will aid in implementing prompt treatment and control measures. The present study standardised LAMP as a point-of-care diagnostic tool to diagnose feline panleukopenia using faecal samples of suspected cats. The assay will reduce the cost and time required for the diagnosis.
Initially, positive cases of feline panleukopenia were identified by PCR amplification of DNA extracted from faecal swabs targeting the VP2 gene of FPV. Amplicons of 698 bp size produced as a result of PCR on sequencing and BLAST revealed 100% similarity to the VP2 gene of FPV. Out of the 120 samples tested, viral DNA was detected in 100 (83.33%) of the suspected samples. Adopting purebred cats as pets is a relatively recent pet parenting practice in the study area (Vincy & Tresamol 2023). The increased occurrence of feline panleukopenia among cats in the study area could be because of a lack of awareness regarding vaccination among cat owners (Mukhopadhyay et al. 2017). The high rate of detection of FPV among cats with suggestive clinical signs indicates an urgent need to create public awareness regarding the vaccination of domestic cats in the study area.
DNA extracted from a known case of FPV formed the template DNA for standardising the LAMP assay (Accession No. https://www.ncbi.nlm.nih.gov/nuccore/%20MN549355.1/). The optimised ratio of inner primer to outer primer was 1:8, magnesium at 16 mM, and betaine at 1.6 mM. The optimal time-temperature combination for successful amplification was 60 °C for 60 minutes. An initial denaturation at 95 °C for five minutes before adding Bst polymerase was required for successful amplification. The primers used in the study had high guanine-to-cytosine (G: C) content, which needs more energy for denaturation, necessitating the initial denaturation step. Better results following initial denaturation for LAMP were reported by Aryan et al. (2010) and Miranda et al. (2017).
Visualisation of the LAMP reaction presents a distinct challenge. The gel electrophoresis of the LAMP product produced a ladder-like pattern with amplicons of different sizes terminating at 200 bp, indicating a successful amplification. These results are consistent with the PCR using outer primers, which also yielded 200 bp amplicon. However, the usefulness of gel electrophoresis is hampered by its reliance on specialised equipment despite its ability to reveal a clear ladder-like structure (Schneider et al. 2019).
Another practical method to visualise the LAMP reaction is to observe the turbidity caused by magnesium pyrophosphate precipitation as the process proceeds. However, appreciation of magnesium pyrophosphate turbidity by the naked eye is difficult when precipitation levels are low (Shirato 2019). The addition of the SYBR Green 1 solution at the end of the LAMP reaction successfully addressed all the above limitations. The solution retained the orange colour if there was no amplification. In instances of positive results, the solution exhibited a definitive transition to an apple-green hue, while in negative cases, the solution retained its original orange colouration. The convenient and unambiguous contrast provided by the SYBR Green 1 method led to its selection as the preferred method for visualising the results in terms of its advantages over the detection of turbidity or agar gel electrophoresis (Lai et al. 2021).
The LAMP assay exhibited a robust detection capability for FPV up to 1.25 ng/μL in the mixed DNA samples extracted from a multicomponent vaccine. While the study utilised a multicomponent vaccine as the positive control DNA, direct quantitation of the virus load in the positive control could have provided more valuable insights into the assay's sensitivity. However, the findings proved that the LAMP developed in this study was highly sensitive and could amplify a low quantity of DNA from clinical samples (Rapichai et al. 2022). The low detection limit of the LAMP test positions it as a superior alternative to newer tests like the SNAP test (Jacobson et al. 2021). The ability to detect FPV at such minimal concentrations enhances diagnostic precision and opens new avenues for early detection and intervention in feline panleukopenia cases. This advancement offers veterinarians a powerful tool to rapidly and accurately identify FPV in clinical samples.
The LAMP assay could not amplify non-target DNA, including E. coli and Felis cattus DNA. The experimental results corresponded well with the in-silico analysis, which predicted that the designed primers had no cross-reactivity with other possible contaminants in feline faecal matter, such as E. coli or other housekeeping genes in cats. However, the test was capable of amplifying canine parvovirus DNA. The close similarity between the VP2 genes of CPV and FPV resulted in non-specific amplification (Decaro et al. 2010; Brindhalakshmi et al. 2016). Mukhopadhyay et al. (2017) reported that both canine parvovirus-2 and FPV circulated in cases of feline panleukopenia in India. Findings corroborate that the test could detect both viruses in clinical settings, emphasising the relevance in point-of-care facilities.
Out of the 120 DNA samples tested, all the 100 PCR-positive samples tested positive in LAMP. Among the 20 PCR-negative samples, 18 were negative in the LAMP assay. However, two PCR-negative samples tested positive in LAMP. The ability of the LAMP assay to detect FPV in PCR-negative samples indicated the increased detection limit of LAMP compared to PCR (Rapichai et al. 2022). Analysis revealed that the LAMP assay was 100% sensitive, 90% specific, and 98.33% accurate compared to PCR, making it an effective diagnostic method in field settings. The LAMP assay could be a reliable tool for point-of-care diagnosis of feline panleukopenia in limited resource settings.
Conclusions
In the present study, a LAMP assay was developed and standardised for the detection of FPV DNA from the faecal samples of cats. The LAMP protocol designed in the present study could detect up to 1.25 ng/μl of the template DNA. The assay could detect feline panleukopenia in 85% of the sample population. Additionally, two PCR-negative samples were found to be LAMP-positive suggesting a better detection limit in the LAMP assay compared to PCR.
Compared to PCR, the sensitivity and specificity of LAMP were found to be 100% and 90%, respectively indicating that LAMP is as sensitive as PCR in detecting FPV from clinical samples. Thus, from the present study, it was concluded that LAMP could be a potential diagnostic tool to detect FPV from clinical samples in laboratories with limited resources and initial investments.
Acknowledgements
The authors acknowledges the support by the Director of Academics and Research, Kerala Veterinary and Animal Sciences University, Dean, College of Veterinary and Animal Sciences Pookode for making this research work possible.
Conflict of interest
The authors declare that they have no competing interests.
Funding source
The funding for this research was entirely utilised from University Research Grant for PG Students by Kerala Veterinary and Animal Sciences University
Ethical approval
Since samples were collected from the animal subjects as a part of routine clinical examinations, ethical approval was not required. However, verbal consent from the owners was taken prior to sample collection and the utmost care was taken to ensure the comfort and wellbeing of the animal subjects while handling them.
ORCID
RA Bakde https://orcid.org/0009-0009-3916-8779
RL Rathish https://orcid.org 0000-0003-2419-4885
L John https://orcid.org 0000-0002-4921 -4996
PM Deepahttps://orcid.org 0000-0002-0945-3589
K Vijayakumar https://orcid.org 0000-0002-0744-2560
References
Aryan, E., Makvandi, M., Farajzadeh, A., et al., 2010, A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex, Microbiol Res 165(3), 21 1-220. https://doi.org/10.1016/j.micres.2009.05.001. [ Links ]
Bergmann, M., Schwertler, S., Speck, S., et al., 2019, Faecal shedding of parvovirus deoxyribonucleic acid following modified live feline panleucopenia virus vaccination in healthy cats, Vet Rec 185(3), 83-83. https://doi.org/10.1136/vr.104661. [ Links ]
Brindhalakshmi, B., Mukhopadhyay, H., Antony, P., et al., 2016, Isolation and molecular characterization of canine and feline parvovirus strains-an updated review, J Dairy Vet Anim Res 3(5), 164-9. https://doi.org/10.15406/jdvar.2016.03.00093. [ Links ]
Chomczynski, P., Mackey, K., Drews, R., et al., 1997, DNAzol®: a reagent for the rapid isolation of genomic DNA, Biotechniques 22(3), 550-553. https://doi.org/10.2144/97223pf01. [ Links ]
Datta, S., Kumar, V., Kumar, V., et al., 2021, Identification of aetiological agents of upper respiratory tract infections in domestic cats, Pharma Innov J SP10(1) 46-48. [ Links ]
Decaro, N., Buonavoglia, D., Desario, C., et al., 2010, Characterisation of canine parvovirus strains isolated from cats with feline panleukopenia, Res Vet Sci 89(2), 275-278. [ Links ]
Hamburger, J., Abbasi, I., Kariuki, C., et al., 2013, Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories, Am J Trop Med Hyg 88(2), 344. https://doi.org/10.4269/ajtmh.2012.12-0208. [ Links ]
Jacobson, L.S., Janke, K.J., Giacinti, J., et al., 2021, Diagnostic testing for feline panleukopenia in a shelter setting: a prospective, observational study, J Feline Med Surg 23(12), 1 192-1 199. https://doi.org/10.1177/1098612X211005301. [ Links ]
Kang, J.-I., Park, N.-Y.,Cho, H.-S., 2006, Detection of canine parvovirus in fecal samples using loop-mediated isothermal amplification, J Vet Diagn Invest 18(1), 81-84. https://doi.org/10.1177/104063870601800111. [ Links ]
Lai, M.Y., Ooi, C.H., Lau, Y.L., 2021, Validation of SYBR green I based closed-tube loop-mediated isothermal amplification (LAMP) assay for diagnosis of knowlesi malaria, Malar J 20(1), 1-6. https://doi.org/10.1186/s12936-021-03707-0. [ Links ]
Miranda, C., Vieira, M., Silva, E., et al., 2017, Genetic analysis of feline panleukopenia virus full-length VP 2 gene in domestic cats between 2006-2008 and 2012-2014, Portugal, Transbound Emerg Dis 64(4), 1 178-1183. https://doi.org/10.1111/tbed.12483. [ Links ]
Mochizuki, M., Horiuchi, M., Hiragi, H., et al., 1996, Isolation of canine parvovirus from a cat manifesting clinical signs of feline panleukopenia, J Clin Microbiol 34(9), 2101. https://doi.org/10.1128/jcm.34.9.2101-2105.1996. [ Links ]
Mukhopadhyay, H.K., Nookala, M., Thangamani, N.R., et al., 2017, Molecular characterisation of parvoviruses from domestic cats reveals emergence of newer variants in India, J Feline Med Surg 19(8), 846-852. https://doi.org/10.1177/1098612X16661375. [ Links ]
Notomi, T., Okayama, H., Masubuchi, H., et al., 2000, Loop-mediated isothermal amplification of DNA, Nucleic Acids Res 28(12), e63-e63. https://doi.org/10.1093/nar/28.12.e63. [ Links ]
Parida, M., Sannarangaiah, S., Dash, P.K., et al., 2008, Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases, Rev Med Virol 18(6), 407-421. https://doi.org/10.1002/rmv.593. [ Links ]
Rapichai, W., Saejung, W., Khumtong, K., et al., 2022, Development of colorimetric reverse transcription loop-mediated isothermal amplification assay for detecting feline coronavirus, Animals 12(16), 2075. https://doi.org/10.3390/ani12162075. [ Links ]
Rehme, T., Hartmann, K., Truyen, U., et al., 2022, Feline panleukopenia outbreaks and risk factors in cats in animal shelters, Viruses 14(6), 1248. https://doi.org/10.3390/v14061248. [ Links ]
Schneider, L., Blakely, H., Tripathi, A., 2019, Mathematical model to reduce loop mediated isothermal amplification (LAMP) false-positive diagnosis, Electrophoresis 40(20), 2706-2717. https://doi.org/10.1002/elps.201900167. [ Links ]
Shirato, K., 2019, Detecting amplicons of loop-mediated isothermal amplification, Microbiol Immunol 63(10), 407-412. https://doi.org/10.1111/1348-0421.12734. [ Links ]
Thrusfield, M., Christley, R., Brown, H. et al., 2018. Veterinary epidemiology. 4th edn, Hoboken, NJ, Wiley. https://doi.org/10.1002/9781118280249. [ Links ]
Vincy, P., Tresamol, P., 2023, Prevalence of gastro-intestinal and haemoparasitic infections among domestic cats of Kerala, J Parasit Dis 1-4. https://doi.org/10.1007/s12639-023-01599-2. [ Links ]
Yang, D.-K., Park, Y.-R., Park, Y., et al., 2022, Isolation and molecular characterization of feline panleukopenia viruses from Korean cats, Korean J Vet Res 62(1), e10. https://doi.org/10.14405/kjvr.20210050. [ Links ]
 Correspondence:
Correspondence:
email: rathish@kvasu.ac.in














