Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Journal of the South African Veterinary Association
On-line version ISSN 2224-9435
Print version ISSN 1019-9128
J. S. Afr. Vet. Assoc. vol.84 n.1 Pretoria Jan. 2013
SHORT COMMUNICATION
Molecular epidemiology of infectious bursal disease virus in Zambia
Christopher J. KasangaI, IV; Tsuyoshi YamaguchiII; Hetron M. Munang'anduIII; Kenji OhyaIV; Hideto FukushiIV
IDepartment of Veterinary Microbiology and Parasitology, Sokoine University of Agriculture, Tanzania
IIAvian Zoonoses Center, Tottori University, Japan
IIIDepartment of Paraclinical Sciences, University of Zambia, Zambia
IVLaboratory of Veterinary Microbiology, Gifu University, Japan
ABSTRACT
Nucleotide sequences of the VP2 hypervariable region (VP2-HVR) of 10 infectious bursal disease viruses detected in indigenous and exotic chickens in Zambia from 2004 to 2005 were determined. Phylogenetic analysis showed that the viruses diverged into two genotypes and belonged to the African very virulent types (VV1 and VV2). In the phylogenetic tree, strains in one genotype clustered in a distinct group and were closely related to some strains isolated in western Africa (VV1), with nucleotide similarities of 95.7% - 96.5%. Strains in the other genotype were clustered within the eastern African VV type (VV2), with nucleotide similarities of 97.3% - 98.5%. Both genotypes were distributed in the southern parts of Zambia and had a unique conserved amino acid substitution at 300 (E→A) in addition to the putative virulence marker at positions 222(A), 242(1), 256(1), 294(1) and 299(S). These findings represent the first documentation of the existence of the African VV-IBDV variants in both indigenous and exotic chickens in Zambia.
Introduction
Infectious bursal disease virus (IBDV) is the causative agent of infectious bursal disease (IBD) of young chickens (Hirai et al. 1974). IBDV is a member of the genus Avibirnavirus of the family Birnaviridae (Chettle, Stuart & Wyeth 1989). The virus infects the IgM-bearing B-lymphocytes in the bursa of Fabricius (Zierenberg et al. 2000), leading to immunosuppression. There are two distinct serotypes of IBDV, namely serotype 1 and serotype 2. IBD is caused by the serotype 1 viruses, which are classified as classical virulent, antigenic variant or very virulent (VV) IBDVs based on their difference in virulence and lethality. Serotype 2 viruses are non-pathogenic to chickens (Hoque et al. 2001). The IBDV genome consists of two segments of double stranded ribonucleic acid (dsRNA). Segment A (3.4 kb) encodes four viral proteins: the two capsid proteins VP2 (48.0 kDa) and VP3 (32.0 kDa - 35.0 kDa), the viral protease VP4 (24.0 kDa) and a nonstructural protein VP5 (17.0 kDa - 21.0 kDa). Segment B (2.8 kb) encodes VP1 (90.0 kDa), an ribonucleic acid (RNA)-dependent RNA polymerase.
The hypervariable region (HVR) within VP2, between amino acid residues 206 and 350, is highly conserved, with the highest amino acid sequence variation amongst serotype 1 strains. The nucleotide and deduced amino acid sequences of this region are widely used for molecular epidemiological studies of IBDV (Kasanga et al. 2007).
Infectious bursal disease virus is worldwide an important virus in the poultry industry as it causes immunosuppression and mortality in infected chickens (Jackwood et al. 2011). The emergence of a pathotypic variant with enhanced virulence, termed very virulent IBDV (VV-IBDV), in Europe in the 1980s led to difficulties in the control of IBD with the use of classical vaccines (Brown, Green & Skinner 1994). The origin of VV-IBDV remains unclear.
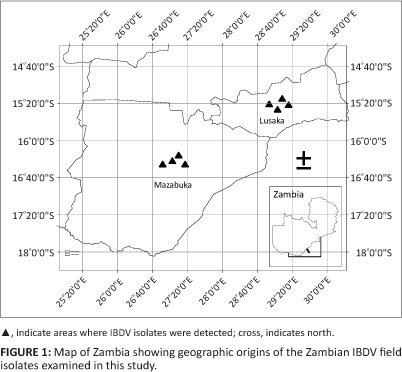
In Zambia, IBD outbreaks have been reported to occur in vaccinated and non-vaccinated chickens from various parts of the country since the early 1990s (Kasanga et al. 2008; Munang'andu, pers. comm., 2005). However, the molecular characteristics of field IBDV isolates from Zambia have not yet been extensively studied, with only a few reports available on IBDV genome reassortment in this country (Kasanga et al. 2012, 2013). The control of IBD in Zambia needs a proper knowledge of the molecular characteristics and antigenicity of the circulating IBDV field strains, as this would help in selecting appropriate vaccine strains for the effective neutralisation of field isolates. In the current study, 10 IBDV field strains obtained from commercial broiler farms and indigenous scavenging chickens collected in Zambia between 2004 and 2005 were characterised (Table 1 and Figure 1).
Materials and methods
The bursae obtained from clinically normal indigenous scavenging chickens and IBD-confirmed dead broiler chickens from different farms were smeared directly onto separate filter papers, fixed with 99% ethanol and transported to Japan for molecular characterisation, as described previously (Kasanga et al. 2008; Maw et al. 2006).Total RNA was isolated from the bursal tissues fixed on filter papers and first-strand complementary DNAs were synthesised as described in a previous report (Kasanga et al. 2008). The VP2-HVRs were amplified by polymerase chain reaction (PCR) using the V1forward primer (5'-CCAGAGTCTACACCATAA-3') and V2 reverse primer (3'-TAGAAAGAGTGGCAACAGG-5') (Yamaguchi et al. 2007). The PCR products were cloned into the plasmid pGEM-T-Easy vector (Promega, Madison WI, USA) and cloned DNAs were sequenced at the Dragon Genomics Center (TAKARA Bio, Mie, Japan) using a Templiphi DNA sequencing Template Amplification Kit, DYEnamic ET dye terminator kit, and MegaBACE1,000 (all Amersham Biosciences, Piscataway NJ, USA). At least five independent clones were sequenced per sample. The nucleotide sequences of 10 field isolates were deposited in the GenBank/ DDBJ/EMBL (accession numbers AB368941 to AB368950 [Table 1]) and analysed with the aid of GENETYXMAC software, version 14.0.1 (GENETYX,Tokyo, Japan).
The nucleotide and deduced amino acid sequences of the Zambian field isolates were aligned using CLUSTAL W (version 1.8.3) and subjected to BLAST searches (http://www.ddbj.nig.ac.jp/search/blast-e.html) to determine their comparison with other strains. The sequences of 107 serotype 1 IBDV isolates showing nearest homology were identified and compared with VP2-HVR sequences of Zambian field strains. The phylogenetic tree for nucleotide sequences was constructed using 42 representative strains, including the Zambian IBDV field isolates. Phylogenetic trees based on VP2-HVR sequences were constructed by the neighbour joining method applying the Kimura two parameter option (Kimura 1980). Topological robustness of the tree was estimated by bootstrap analysis of 1000 replicates (Figure 2). The deduced amino acid sequences of 10 Zambian strains were aligned and compared to those of selected serotype 1 IBDVs, including those of classical strains, antigenic variants and VV-IBDVs.
Results and discussion
In a phylogenetic tree based on the nucleotide sequences of VP2-HVRs, all Zambian strains fell within the African VV type (Figure 2). In the tree, the African VV type is subdivided into two major clusters, tentatively named VV1 and VV2. VV1 and VV2 contain strains derived from Nigeria in West Africa and Tanzania in East Africa, respectively. A third cluster, VV3, contains the European VV strains derived from different continents (including Africa, Europe, Asia and elsewhere).
In the VV1 cluster, four Zambian strains were grouped together and formed a distinct subcluster, VV1-Z. The Zambian strains in subcluster VV1-Z were genetically closely related to the Nigerian strains in VV1 (Figure 2). The Zambian strains in VV1-Z showed 97.3% - 98.5% nucleotide sequence identities compared with the VV-IBDVs clustered in VV1, which were isolated from Nigeria. All four Zambian strains in subcluster VV1-Z were isolated from broiler chickens.
The close phylogenetic relationship of VV1-Z to other viruses in VV1 viruses indicates that the strains in the VV1-Z subcluster were derived from a common ancient ancestor of the VV1 groups, with VV1-Z viruses having evolved independently in Zambia. Zambia and Nigeria, located in southern and central, and west Africa, respectively, are geographically distant from each other. Detecting VV1-clustered viruses in Zambia that are closely related to those in Nigeria suggests that similar viruses clustered in VV1 may exist in other countries in Africa.
The VV2 cluster consisted of two distinct subclusters, VV2-Z and VV2-T. Six Zambian strains were grouped in the VV2-Z subcluster (Figure 2). The Zambian strains in VV2-Z showed nucleotide sequence identities of 95.7% -96.5% correspondence to the Tanzanian VV-IBDVs clustered in VV2-T. Of these, four strains (KZC-7, KZC-10, KZC-31 and KZC-113) were isolated from indigenous scavenging chickens, whilst the other two (KZC-107 and KZC-109) were isolated from exotic broiler chickens. The existence of two distinct subclusters in VV2 indicates that the viruses are closely related and share the same ancestor but have undergone independent evolution in different geographic locations (Zambia and Tanzania), leading to segregation of the subclusters.
It has been documented that the viruses in the VV2-T subcluster were isolated from both exotic and indigenous chickens in Tanzania (Kasanga et al. 2007). Previous observations and the current findings suggest that several factors other than the virus host could influence the pattern of virus evolution in Zambia and Tanzania, resulting in two different virus lineages.
All 10 Zambian isolates conserved putative virulence marker amino acids at positions 222(A), 242(I), 256(I), 294(I) and 299(S), which have been identified in most VV-IBDVs (Kasanga et al. 2007). In addition, they retained the amino acid substitution at 300 (E→A) in the minor hydrophilic region of the VP2-HVR domain, as was also observed in the African VV-IBDVs from Tanzania (Kasanga et al. 2007). The conservation of amino acid substitutions in the hydrophilic region implies that the Zambian strains are antigenically different from classical and European VV-IBDVs. It is hypothesised that the amino acid substitution at 300 (E→A) could be associated with the altered biological characteristics of viruses in Zambia. Further reverse genetics studies are recommended to elucidate the role of amino acid substitution at 300 (E→A) in the pathogenicity and lethality of the Zambian field IBDV strains.
Conclusion
In summary, 10 Zambian field IBDV isolates were characterised. It was found that the African VV-IBDVs exist in Zambia and could be divided into two clusters. For in-depth evolutionary analysis of the African VV-IBDVs detected in Zambia, genome segment B of the viruses also needs to be studied in order to provide relevant information required for the design of appropriate control method(s) for emergent African VV-IBDVs.
Acknowledgements
We wish to thank participating farmers in Zambia. This study was supported by Grants-in-Aid for Basic Scientific Research, number (A) 17255010 and (C) 18580308 from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Publication of this article was sponsored by the South African Poultry Association.
Competing interests
The authors declare that they have no financial or personal relationships that may have inappropriately influenced them in writing this article.
Authors' contributions
C.J.K. (Sokoine University of Agriculture) participated in the study design, the experimental work, the analysis and interpretation of the data, and drafted the manuscript. T.Y. (Tottori University), K.O. (Gifu University) and H.F. (Gifu University) participated in the study design, the experimental work and drafting the manuscript. H.M.M. (University of Zambia) participated in sample collection, the experimental work and writing of the manuscript. All authors read and approved the final manuscript.
References
Brown, M.D., Green, P. & Skinner, M.A., 1994, 'VP2 sequences of recent European 'very virulent' isolates of infectious bursal disease virus are closely related to each other but are distinct from those of 'classical' strains', Journal of General Virology 75, 675-680. http://dx.doi.org/10.1099/0022-1317-75-3-675, PMid:8126466 [ Links ]
Chettle, N., Stuart, J.C. & Wyeth, P.J., 1989,'Outbreak of virulent infectious bursal disease in East Anglia', Veterinary Record 125, 271-272. http://dx.doi.org/10.1136/vr.125.10.271, PMid:2552640 [ Links ]
Hirai, K., Shimakura, S., Kawamoto, E., Taguchi, F., Kim, S.T., Chang, C.N. et al., 1974, 'The immunodepressive effect of infectious bursal disease virus in chickens', Avian Diseases 18, 50-57. http://dx.doi.org/10.2307/1589241, PMid:4360708 [ Links ]
Hoque, M.M., Omar, A.R., Chong, L.K., Hair-Bejo, M. & Aini, I., 2001, 'Pathogenicity of Sspl-positive infectious bursal disease virus and molecular characterization of the VP2 hypervariable region', Avian Pathology 30, 369-380. http://dx.doi.org/10.1080/03079450120066377, PMid:19184922 [ Links ]
Jackwood, D.J., Sommer-Wagner, S.E., Crossley, B.M., Stoute, S.T., Woolcock, P.R. & Charlton, B.R., 2011, 'Identification and pathogenicity of a natural reassortant between a very virulent serotype 1 infectious bursal disease virus (IBDV) and a serotype 2 IBDV', Virology 420, 98-105. http://dx.doi.org/10.1016/j.virol.2011.08.023, PMid:21955938 [ Links ]
Kasanga, C.J., Yamaguchi, T., Munang'andu, H.M., Ohya, K. & Fukushi, H., 2013, 'Genomic sequence of an infectious bursal disease virus isolate from Zambia: classical attenuated segment B reassortment in nature with existing very virulent segment A', Archives of Virology 158, 685-689. http://dx.doi.org/10.1007/s00705-012-1531-4, PMid:23129132 [ Links ]
Kasanga, C.J., Yamaguchi, T., Munang'andu, H.M., Wambura, P.N., Ohya, K. & Fukushi, H., 2012, 'Genomic sequence of infectious bursal disease virus from Zambia suggests evidence for genome re-assortment in nature', Onderstepoort Journal of Veterinary Research 79, E1. http://dx.doi.org/10.4102/ojvr.v79i2.473 [ Links ]
Kasanga, C.J., Yamaguchi, T., Wambura, P.N., Maeda-Machang'u, A.D., Ohya, K. & Fukushi, H., 2007, 'Molecular characterization of infectious bursal disease virus (IBDV): diversity of very virulent IBDV in Tanzania', Archives of Virology 152, 783790. http://dx.doi.org/10.1007/s00705-006-0898-5, PMid:17226068 [ Links ]
Kasanga, C.J., Yamaguchi, T., Wambura, P.N., Munang'andu, H.M., Ohya, K. & Fukushi, H., 2008, 'Detection of infectious bursal disease virus (IBDV) genome in free-living pigeon and guinea fowl in Africa suggests involvement of wild birds in the epidemiology of IBDV', Virus Genes 36, 521-529. http://dx.doi.org/10.1007/s11262-008-0219-z, PMid:18343984 [ Links ]
Kimura, M., 1980, 'A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences', Journal of Molecular Evolution 16, 111-120. http://dx.doi.org/10.1007/BF01731581, PMid:7463489 [ Links ]
Maw, M.T., Yamaguchi, T., Kasanga, C.J., Terasaki, K. & Fukushi, H., 2006, 'A practical tissue sampling method using ordinary paper for molecular detection of infectious bursal disease virus RNA by RT-PCR', Avian Diseases 50, 556-560. http://dx.doi.org/10.1637/7537-032806R.1, PMid:17274294 [ Links ]
Yamaguchi, T., Kasanga, C.J., Terasaki, K., Maw, M.T., Ohya, K. & Fukushi, H., 2007, 'Nucleotide sequence analysis of VP2 hypervariable domain of infectious bursal disease virus detected in Japan from 1993 to 2004', Journal of Veterinary and Medical Sciences 69, 733-738. http://dx.doi.org/10.1292/jvms.69.733, PMid:17675805 [ Links ]
Zierenberg, K., Nieper, H., Van den Berg, T.P., Ezeokoli, C.D., Voss, M. & Muller, H., 2000, 'The VP2 variable region of African and German isolates of infectious bursal disease virus: comparison with very virulent, "classical" virulent, and attenuated tissue culture-adapted strains', Archives of Virology 145, 113-125. http://dx.doi.org/10.1007/s007050050009, PMid:10664410 [ Links ]
 Correspondence:
Correspondence:
Christopher Kasanga
PO Box 3019
Chuo Kikuu, Morogoro, Tanzania
Email: chrisskasa@gmail.com
Received: 24 July 2012
Accepted: 01 July 2013
Published: 28 Oct. 2013














