Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
Journal of the South African Veterinary Association
versão On-line ISSN 2224-9435
versão impressa ISSN 1019-9128
J. S. Afr. Vet. Assoc. vol.81 no.4 Pretoria Dez. 2010
ARTICLE ARTIKEL
Prevalence of mixed Trypanosoma congolense infections in livestock and tsetse in KwaZulu-Natal, South Africa
K Gillingwater; M V Mamabolo; P A O Majiwa*
Agricultural Research Council, Onderstepoort Veterinary Institute, Private Bag X05, Onderstepoort, 0110 South Africa
ABSTRACT
Trypanosoma congolense causes the most economically important animal trypanosomosis in Africa. In South Africa, a rinderpest pandemic of the 1890s removed many host animals, resulting in the near-eradication of most tsetse species. Further suppression was achieved through spraying with dichlorodiphenyltrichloroethane (DDT); however, residual populations of Glossina austeni and G. brevipalpis remained in isolated pockets. A total of 506 of these tsetse flies were captured in the Hluhluwe-iMfolozi Park, the St Lucia Wetland Park and Boomerang commercial farm. The polymerase chain reaction (PCR) was used to determine the infection rate and frequency of mixed infections of these flies. Additionally, 473 blood samples were collected from cattle at communal diptanks and a commercial farm in the area and each one examined by the haematocrit centrifugation technique (HCT). Furthermore, buffy coats from these blood samples were spotted onto FTA Elute cards and the DNA extracted from each one tested using 3 separate PCRs. The HCT revealed the presence of trypanosomes in only 6.6 % of the blood samples; by contrast, species-specific PCR detected trypanosome DNA in 50 % of the samples. The species-specific PCR detected trypanosome DNA in 17 % of the tsetse flies, compared with the nested PCR targeting rDNA which detected trypanosome DNA in only 14 % of the samples. Over time, the transmission of Savannah-type T. congolense and Kilifi-type T. congolense as mixed infections could have an impact on disease manifestation in different hosts in the area.
Keywords: mixed infection, PCR, prevalence, Trypanosoma congolense, tsetse.
INTRODUCTION
Trypanosoma congolense, although the smallest of the trypanosome species, remains the most pathogenic to animals2. Tsetse-transmitted trypanosomes infective to livestock cause huge economic losses to the livestock industry18 and tsetse flies (Glossina spp.) currently infest over 10 million square kilometres of fertile land distributed among 37 countries within the African continent.
South Africa was fairly successful at controlling the spread of nagana; sleeping sickness, the equivalent disease in humans, has not been observed in the country. The tsetse populations declined drastically after rinderpest swept through the country in the 1890s and, combined with aggressive spraying with DDT, large areas such as the Kruger National Park were freed of the fly. Unfortunately, small isolated populations of tsetse survived in several game reserves in the northeastern parts of the then Zululand (now part of KwaZulu-Natal Province (KZN)) and by 1905, nagana was again a major threat to animal health3. Once infected, domestic and wild animals can remain so for life, providing a constant reservoir of the parasite.
Several outbreaks of nagana, due to T. congolense infections, were reported in South Africa until 1945. It was not until 1990, however, when a series of widespread outbreaks of nagana occurred, that G. brevipalpis and G. austeni were established to be responsible3. These 2 species of tsetse are now found in small isolated pockets in KwaZulu-Natal7. Additionally there are fears that G. morsitans may re-invade the Kruger National Park through the recently established Great Limpopo Transfrontier Park (www.greatlimpopopark.com).
Preliminary surveys into the recent situation of trypanosomoses in the Hluhluwe-iMfolozi Park, KwaZulu-Natal, indicated that the majority of cases were due to Savannah-type T. congolense20. Recently, Kilifi-type T. congolense was found in cattle and tsetse near this game park12. Furthermore, a mixed infection of both Savannah-type T. congolense and Kilifi-type T. congolense was discovered12. Within the group of trypanosomes classified as T. congolense, 5 different genotypes have so far been found using various DNA probes6, 10. Although these trypanosomes occur as mixed infections in different tsetse vectors and animal hosts, little information exists on the significance of the mixed infections. Such infections may have an impact on smallholder livestock farming near game parks. This study was therefore conducted to investigate more thoroughly the frequency of mixed infections of T. congolense genotypes in tsetse flies and cattle, and the possible significance this could have on livestock farming and game reserves in South Africa. The study sought to determine the trypanosome infections present in tsetse flies and cattle at specific locations in the Hluhluwe-Umfolozi region in KwaZulu-Natal and to verify the frequency of mixed infections and the infection rate of trypanosomes in both tsetse flies and cattle.
MATERIALS AND METHODS
Field site
All fieldwork was conducted in and around the Hluhluwe-iMfolozi area in KwaZulu-Natal, South Africa. The exact locations of the communal diptanks and the commercial cattle farm sampled have been previously described12, except that of Ndabendabe, which is equidistant to the north of Ocilwane and to the south of Mvutshini diptanks. Microscopy on, and blotting of, samples was done at the ARC Tsetse Research Station, Kuleni, KZN; all the subsequent analyses were performed at the ARC-OVI, Onderstepoort.
Collection of blood samples from cattle
Fresh venous blood was collected from cattle at 4 communal diptanks (Mvutshini, Ocilwane, Ekuphindisweni and Ndabendabe) and from cattle on a commercial farm (Boomerang). The cattle were randomly sampled and included both weaners and adults of Nguni and Brahman breeds. Blood was taken from the median coccygeal vein using 10 mℓ EDTA-coated Vacutainer® tubes (Becton Dickinson, Plymouth, UK), labelled and then placed on ice in a cooler box for transportation to the Tsetse Research Station, Kuleni.
Collection of tsetse flies
Tsetse flies were captured using the 'Horizontal-' or 'H-trap' baited with synthetic ox-odour (4-methyl phenol, acetone and octanol)7. Traps were set up at Boomerang (a commercial cattle farm), at Charter's Creek (St Lucia Greater Wetland Park) and in the Hluhluwe-iMfolozi Park. They were inspected in the morning (06:00) and in the afternoon (18:00) for captured flies. Bottles containing trapped flies were labelled with location, trap number, date and time and then replaced with empty ones. The bottles with flies were packed in a polystyrene box, covered with a damp cloth to provide a humidified atmosphere and transported to the Tsetse Research Station, Kuleni.
Sample preparation
In the laboratory at the station, blood from the cattle samples was placed into heparinised capillaries, stoppered at one end with plasticine and spun in a haematocrit centrifuge at 12000 rpm for 10 min. The buffy coat that formed was then examined using a light microscope under ×40 magnification for the presence of trypanosomes. In addition, packed cell volume (PCV) of each animal sampled was recorded. Thereafter, capillaries were cut using a diamond cutter and the buffy coat layer (approximately 40 µℓ) spotted onto FTA Elute filter cards (Whatman, Biosciences Ltd, Brentford, UK). Each circle on a card represented 1 animal sampled. The cards were then labelled and allowed to dry thoroughly before storage.
Captured tsetse flies were placed into Petri-dishes and then on ice for 10-15 min to render them inactive. Data (location, trap number, date, time, sex and species) for each fly was recorded, before the midgut and proboscis were separately dissected, examined under light microscopy for the presence of trypanosomes and then placed on separate FTA Elute filter cards, together with 20 µℓ phosphate-buffered saline with glucose (PSG). Each circle on a card represented 1 tsetse fly, with midguts and proboscis each placed separately for DNA analysis. Fly midguts and proboscis were placed on different sets of FTA Elute filter cards to avoid contamination between them. The cards were then labelled and allowed to dry thoroughly before storage. All utensils used during fly dissection were cleaned with 70 % ethanol between both midgut and proboscis extraction and again between each fly.
DNA elution
For all the samples, DNA was eluted from the FTA Elute filter cards according to the manufacturer's instructions. Briefly, a 3-mm-disk was punched out of each circle and placed into a 1.5 mℓ reaction tube containing 500 µℓ of nucleasefree water. These were subjected to pulse mixing before being transferred to a new 0.5 mℓ reaction tube containing 30 µℓ of nuclease-free water, then onto a heating block and incubated at 95 ºC for 30 min. Thereafter, the samples were spun at 13 000 rpm (Heraeus Biofuge pico, DJB Labware, UK) for 30 s and each disk was gently removed using sterile pipette tips. The eluted DNA was then stored at -20 ºC until further use.
Amplification reactions
Trypanosome DNA was detected employing 2 different assays, 1 using 2 separate amplification reactions, targeting a segment of the 18S ribosomal RNA gene in nested reactions5 and the other the species-specific DNA sequences11, 14. The 1st amplification reaction used the primer set 18ST nF2 (CAA CGA TGA CAC CCA TGA ATT GGG GA) and 18ST nR3 (TGC GCG ACC AAT AAT TGC AAT AC). The product of this was then committed to the nested reaction using the primers 18ST nF2 and 18ST nR2 (GTG TCT TGT TCT CAC TGA CAT TGT AGT G). Both reactions were carried out in a final volume of 25 µℓ , containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 200 µM of each dNTP, 0.8 µM of each of the primers and 0.5 U ExTaq polymerase (Takara Bio Inc.) and 2.5 µℓ of the eluted DNA or, for the nested reaction, 0.5 µℓ of products from the 1st PCR. The thermocycling parameters have been previously described5.In all reactions, DNA from T. congolense IL300019 and T. congolense 70989 were included as positive controls for Savannah-type and Kilifi-type T. congolense, respectively. Master reaction mix containing no DNA served as a negative control.
For the species-specific PCR, the following primer set was used to detect Savannah-type T. congolense, TCN1 (TCG AGC GAG AAC GGG CAC TTT GCG A) and TCN2 (ATT AGG GAC AAA CAA ATC CCG CAC)11 and the primer set TCK1 (GTG CCC AAA TTT GAA GTG AT) and TCK2 (ACT CAA AAT CGT GCA CCT CG)14 was used to detect Kilifi-type T. congolense. All species-specific PCRs were performed in a final volume of 25 µℓ , containing 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 100 µM of each dNTP, 0.4 µM of each of the primers, 0.5 U ExTaq polymerase (Takara Bio Inc.) and 2.5 µℓ of the eluted DNA. The thermocycling parameters were set at 94 ºC for 1 min for initial denaturation, followed by 30 cycles of 30 s at 94 ºC, 30 s at 60 ºC and 1 min at 72 ºC.
All PCR products obtained from the amplification reactions were visualised by electrophoresis in 2 % agarose gels stained with ethidium bromide, which was then photographed.
RESULTS
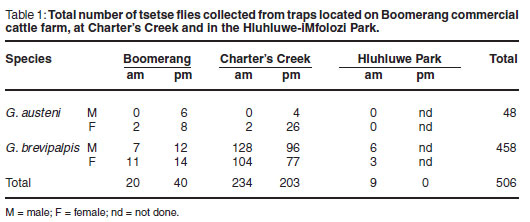
In all, 506 tsetse flies were collected and sampled; 60 at Boomerang, 437 at Charter's Creek and 9 at Hluhluwe-iMfolozi Park (Table 1). The majority of the flies were G. brevipalpis caught overnight, while G. austeni were caught mainly during the day. No G. austeni were caught in the Hluhluwe-iMfolozi Park at the time of sampling.
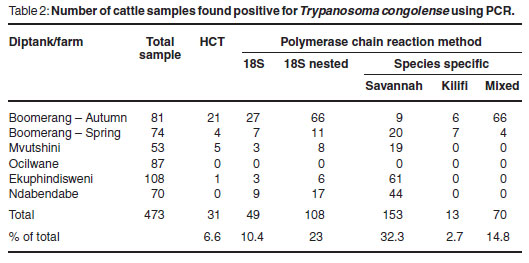
In total, 473 samples were collected from cattle and examined using the HCT and PCR targeting the 18S ribosomal RNA gene locus and the species-specific DNA sequences. Overall, 6.6 % (31/473) of the cattle sampled were found to be parasitologically positive by the HCT. The species-specific PCR detected the greatest number of positive samples, 166 out of 473, when compared with the other assay targeting the 18S rRNA gene locus. The overall results obtained for the samples from cattle are shown in Table 2, together with the percentage of cattle sampled which were found to be positive. Assuming that presence of trypanosome DNA in buffy coat is indicative of infection, an average infection rate of 39 % was observed for cattle sampled at these communal diptanks, all of which were attributed to single infections with Savannahtype T. congolense. For Boomerang commercial farm, an average infection rate of 71 % was observed for cattle sampled. The PCV values of the cattle sampled at the communal diptanks ranged from 16-34% for Mvutshini, 10-36% for Ocilwane, 19-32% for Ekuphindisweni and 16-34% for Ndabendabe. Generally, the cattle had low PCV values, appeared emaciated and were in bad condition. The PCV values of the cattle sampled at Boomerang commercial farm ranged from 17-40 % in autumn and 18-40 % in spring.
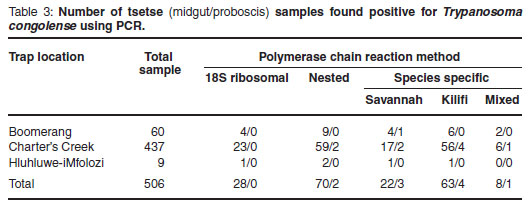
The 506 tsetse flies collected were also examined using PCR, with the speciesspecific PCR detecting the greatest number of positive samples in either the midgut or proboscis (Table 3). For the tsetse midgut samples investigated (Table 3), the percentage of positive infections detected at Boomerang commercial farm was 20 %, compared with the percentage of positive infections detected at Charter's Creek which was 18 %. The percentage positive infections detected at the Hluhluwe-iMfolozi Park was 22 %. No mixed infections with both Savannahand Kilifi-type T. congolense were found in the samples collected from Hluhluwe-iMfolozi Park. In general, an average tsetse midgut infection rate of 20 % was observed. Similarly for the tsetse proboscis samples investigated (Table 3), the percentage of positive infections detected at Boomerang commercial farm and at Charter's Creek was 1.6 %. No positive proboscis infections were detected in the Hluhluwe-iMfolozi Park. In general, an average tsetse proboscis infection rate of 1.6 % was observed.
DISCUSSION
This study was conducted to determine the frequency of trypanosome infections of cattle and tsetse flies at specific locations in South Africa and to initiate an investigation into the prevalence of mixed infections comprising 2 different genotypic groups of T. congolense. Both have a bearing on livestock farming next to game reserves in South Africa.
Tsetse flies collected in the study area were predominantly G. brevipalpis, captured during early dawn and at dusk, compared with G. austeni, which were less abundant and appeared most active during daylight hours. Both males and females of the tsetse species were captured in this study, yet approximately 4 times more females than males G. austeni were collected. As explained in a previous study4, the Index of Apparent Abundance (IAA) of G. brevipalpis was found to be substantially higher than that of G. austeni. Since the H-traps used were specifically designed to capture these 2 Glossina species8, the data collected from this study support the findings of previous studies on tsetse performed in this area of KwaZulu-Natal4, 7, 8. G. brevipalpis is now widely distributed and found in dense indigenous forest, exotic plantations and open grassland. G. austeni on the other hand is associated only with dense forest, enabling it to be close to its preferred hosts (bushpigs and duikers22). G. austeni is currently considered the more important vector involved in transmission of animal trypanosomoses; however, with such high numbers of G. brevipalpis being more widely distributed in this area, the impact that this species may have on disease transmission requires more detailed investigation.
The PCR consistently detected trypanosomes in more cases than did micros-copy13 and the species-specific PCR detected the greatest number of positive samples in both cattle and tsetse. Of the 473 samples collected from cattle in the study area, only 31 (6.6 %) had trypanosomes detectable using the HCT. An explanation for the lower detection of parasite-positive cattle observed during the spring sampling session (September to November) in comparison with the autumn sampling session (March to May) could be attributed to seasonal variation in the number of tsetse vectors available to transmit the disease. During the autumn months, tsetse flies are more abundant and have a higher probability of having fed upon other infected animals, ingesting the parasite and transmitting it to new hosts. In spring, many tsetse flies are still emerging from the larval stages and have yet to obtain their 1st blood meal. The situation at Boomerang commercial farm was further complicated in that trypanocidal drug treatment had been applied to the cattle several months before the spring sampling occurred and, while many parasites may have been cleared from the bloodstream, trypanosome DNA may still have been present, hence providing the higher number of PCR-positive results observed. A similar reason could explain in part why no cattle sampled at Ndabendabe were found positive by HCT, yet were positive by PCR. Since Ndabendabe was included in the study design for the 1st time here, there are no previous data available for comparison and little is known about the history of the trypanocidal drug treatment of cattle in this area.
A recorded 20 % of tsetse flies collected had midgut infections of which 1.6 % were mature. When trypanosomes are found within the tsetse proboscis, such an infection is considered mature, since the fly can transmit the parasite to a host upon which it feeds. The percentage of mature infections found in wild tsetse is usually extremely low (less than 1 %), which is in agreement with a 1.6 % proboscis infection rate observed in this study.
Mixed infections found in the tsetse (both midgut and proboscis) could have occurred through several possible sources. Firstly, the tsetse may have fed on a host infected with only Savannah-type T. congolense and subsequently on another host infected with only Kilifi-type T. congolense. This would have provided a midgut infection, which may later have developed into a mature infection consisting of a mixture of the 2 genotypic groups. Alternatively, the tsetse may have fed on a host with a pre-established mixed infection consisting of both Savannahtype T. congolense and Kilifi-type T. congolense.
Mating apparently occurs in T. congolense16, although it is not obligatory17. Whether any of the genetically distinct groups of T. congolense, such as the Kilifiand Savannah-type, can mate, remains unknown. When equal numbers of a clonal population of Savannah-type T. congolense and a clonal population of Kilifi-type T. congolense are experimentally injected into mice, the parasitaemia which develops does not contain equal numbers of the 2 (data not shown). In fact, Savannah-type T. congolense grows rapidly, producing much higher parasitaemia than Kilifi-type T. congolense. The mice survive for longer than when infected with only Savannah-type T. congolense, demonstrated normally as an acute infection, where mortality occurs within several weeks post-infection. Mice infected with Kilifi-type T. congolense often display a chronic infection, where an initial wave of parasitaemia can be detected, but thereafter parasitaemia disappears and the mice remain chronically infected.
Several possible scenarios could explain this observation. The mixed infection between the 2 T. congolense genotypes may lead to the formation of recombinant trypanosomes, which share qualities of both parental genotypes, thus explaining the appearance of a less acute disease progression in the mice, yet still virulent enough to eventually cause mice to succumb to infection. However, there is currently no evidence of whether mating can occur between genetically distinct groups of T. congolense. A 2nd possible explanation is competition between the 2 genotypes present within the mice, implying that factors influencing the virulence of the 2 trypanosome genotypes may play a vital role. It has been demonstrated that genetically different T. congolense strains belonging to the same genetic subgroup taken from the same host in 1 geographical area can differ substantially in their levels of virulence, thus leading to differences in disease manifestation15. Studies have demonstrated that both intra-specific competition and mutual competitive suppression occur among trypanosomes in a mixed infection1, both leading to prolonged host survival. Strength of suppression appears to be dependant on strain density and host survival is prolonged when a less virulent strain is present in the infection. If infection with Kilifi-type T. congolense in mice (or in cattle) presents prolonged host survival and co-infection with Savannah-type T. congolense occurs, causing mutual suppression within that host, then this can have great implications for management of nagana due to T. congolense.
Many wildlife species are trypanotolerant and act as reservoirs of nagana. With the migration of people and their livestock into or near areas inhabited by wildlife species, in search of arable farmland and pastures, settings emerge which present tsetse flies with another source of food, enabling new transmission cycles to form from sylvatic to domestic21. Farmers grazing their cattle close to wildlife reserves and game parks place susceptible livestock at greater risk of being infected with diseases in wildlife. The impact of nagana on susceptible livestock is likely to change due to environmental alterations and these will have to be considered when planning control strategies against the disease.
ACKNOWLEDGEMENTS
The Department of Agriculture, Forestry and Fisheries, South Africa, provided funds for this study. K. Gillingwater was supported by the Swiss National Science Foundation (SNSF), Switzerland, through a research grant (Grant number PBBSB120874). We thank the staff at the ARC Tsetse Research Station, Kuleni for their assistance and the farmers who brought their cattle to the diptanks for allowing us to sample the cattle. We thank also our colleagues Makhosazana Motloang and Nomsa Letsoalo for their support, and Dr Marco Romito and Mr Arthur Spickett for critical reading of the manuscript.
REFERENCES
1. Balmer O, Stearns S C, Schötzau A, Brun R 2009 Intraspecific competition between co-infecting parasite strains enhances host survival in African trypanosomes. Ecology 90: 3367-3378 [ Links ]
2. Bengaly Z, Sidibe I, Ganaba R, Desquesnes M, Boly H, Sawadogo L 2002 Comparative pathogenicity of 3 genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Veterinary Parasitology 108: 1-19 [ Links ]
3. Bosman P P 1990 Trypanosomosis (Trypanosoma congolense) in South Africa. Office International des Epizooties: Disease Information 3: 79-80 [ Links ]
4. Esterhuizen J, Kappmeier Green K, Marcotty T, Van den Bossche P 2005 Abundance and distribution of the tsetse flies, Glossina austeni and G. brevipalpis, in different habitats in South Africa. Medical and Veterinary Entomology 19: 367-371 [ Links ]
5. Geysen D, Delespaux V, Geerts S 2003 PCR-RFLP using Ssu-rDNA amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Veterinary Parasitology 110: 171-180 [ Links ]
6. Gibson W 2009 Species-specific probes for the identification of the African tsetsetransmitted trypanosomes. Parasitology 136: 1501-1507 [ Links ]
7. Kappmeier K 2000 A newly developed odour-baited 'H trap' for the live collection of Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in South Africa.Onderstepoort Journal of Veterinary Research 67: 15-26 [ Links ]
8. Kappmeier K Nevill E M 1999 Evaluation of a proposed odour-baited target to control the tsetse flies Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in South Africa. Onderstepoort Journal of Veterinary Research 66: 327-332 [ Links ]
9. Ledoka M V 2008 Molecular characterization of trypanosomes commonly found in cattle, wild animals and tsetse flies in KwaZulu-Natal, South Africa 2005-2007. MSc thesis, University of Pretoria [ Links ]
10. Majiwa P A O, Maina M, Waitumbi J N, Mihok S, Zweygarth E 1993 Trypanosoma (Nannomonas) congolense: molecular characterization of a new genotype from Tsavo, Kenya. Parasitology 106: 151-162 [ Links ]
11. Majiwa P A O, Thatthi R, Moloo S K, Nyeko J P H, Otieno L H, Maloo S 1994 Detection of trypanosome infections in the saliva of tsetse flies and buffy coat samples from antigenaemic but aparasitaemic cattle. Parasitology 108: 1-10 [ Links ]
12. Mamabolo M V, Ntantiso L, Latif A, Majiwa P A O 2009 Natural infection of cattle and tsetse flies in South Africa with 2 genotypic groups of Trypanosoma congolense. Parasitology 136: 425-431 [ Links ]
13. Marcotty T, Simukoko H, Berkvens D, Vercruysse J, Praet N, Van den Bossche P 2008 The use of the PCV-value in the diagnosis of trypanosomal infections in cattle. Preventive Veterinary Medicine 87: 288-300 [ Links ]
14. Masiga D K, Smyth A J, Hayes P, Bromidge T J, Gibson W C 1992 Sensitive detection of trypanosomes in tsetse flies by DNA amplification. International Journal of Parasitology 22: 909-918 [ Links ]
15. Masumu J, Marcotty T, Geysen D, Geerts S, Vercruysse J, Dorny P, Van den Bossche P 2006 Comparison of the virulence of Trypanosoma congolense strains isolated from cattle in a trypanosomiasis endemic area of eastern Zambia. International Journal for Parasitology 36: 497-501 [ Links ]
16. Morrison L J, Tweedie A, Black A, Pinchbeck G L, Christley R M, Schoenefeld A, Hertz-Fowler C, MacLeod A, TurnerCMR, Tait A 2009. Discovery of mating in the major African livestock pathogen Trypanosoma congolense. PLoS One 4: e5564 [ Links ]
17. Peacock L, Ferris V, Bailey M, Gibson W 2009. Intraclonal mating occurs during tsetse transmission of Trypanosoma brucei. Parasites and Vectors 21: 43. [ Links ]
18. Shaw A P 2009 Assessing the economics of animal trypanosomosis in Africa-history and current perspectives. Onderstepoort Journal of Veterinary Research 76: 27-32 [ Links ]
19. Urakawa T, MajiwaPAO 2001 Physical and transcriptional organization of the ribosomal RNA genes of the savannah-type Trypanosoma congolense. Parasitology Research 87: 431-438 [ Links ]
20. Van den Bossche P, Esterhuizen J, Nkuna R, Matjila T, Penzhorn B, Geerts S, Marcotty T 2006. An update of the bovine trypanosomosis situation at the edge of HluhluweiMfolozi Park, KwaZulu-Natal Province, South Africa. Onderstepoort Journal of Veterinary Research 73: 77-79 [ Links ]
21. Van den Bossche P, de La Rocque S, Hendrickx G, Bouyer J 2010 A changing environment and the epidemiology of tsetse-transmitted livestock trypanosomiasis. Trends in Parasitology 26: 236-243 [ Links ]
22. Weitz B 1963. The feeding habits of Glossina. Bulletin of the World Health Organization 28: 711-729 [ Links ]
Received: August 2010.
Accepted: November 2010.
* Author for correspondence. E-mail: majiwap@arc.agric.za














