Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
Journal of the South African Veterinary Association
On-line version ISSN 2224-9435
Print version ISSN 1019-9128
J. S. Afr. Vet. Assoc. vol.81 n.3 Pretoria Sep. 2010
ARTICLE ARTIKEL
Prevalent organisms on ostrich carcasses found in a commercial abattoir
L C HoffmanI,*; T J BritzII; D C SchnetlerII
IDepartment of Animal Sciences, University of Stellen bosch, Private Bag X1, Matieland, 7602 South Africa
IIDepartment of Food Science, Stellenbosch University, Private Bag X1, Matieland, 7602 South Africa
ABSTRACT
The prevalent microbial growth on carcasses before and after overnight cooling in an ostrich abattoir and de-boning plant was investigated. The effect of warm or cold trimming of the carcasses was examined together with possible causes of contamination along the processing line. An attempt was made to link the prevalent microorganisms that were identified from carcasses to those from specific external contamination sources. Samples of carcasses and possible contaminants were collected in the plant, plated out and selected organisms were typed using a commercial rapid identification system. It was indicated that the cold trim (mainly of bruises) of carcasses was advantageous in terms of microbiological meat quality. Results indicated pooled water in the abattoir as the most hazardous vector for carcass contamination and that contaminants from this source are mostly Gram-negative pathogens. Pseudomonas and Shigella were frequently isolated from surface and air samples and indicated that the control of total plant hygiene is a requirement for producing ostrich meat that is safe to consume and has an acceptable shelf-life.
Keywords: cold trimming, contamination, microbial growth, warm trimming.
INTRODUCTION
The meat of animal carcasses is sterile while protected by the skin or by the hide10. In most instances, immediately after slaughter, the skins/hides are removed and the meat is exposed. From the moment of the 1st incision in the skin during the flaying process, bacterial contamination may occur3. Microbial contamination can come from a wide range of external sources, including workers' hands and clothing, water supply, air supply and the slaughter equipment. These contaminating organisms can cause bacterial spoilage of the meat, loss of shelf-life in the end product or in the worst case, food infection or poisoning of the consumers. Throughout the abattoir or processing plant these vectors of microbiological contamination should be identified and monitored to minimise the damage done to the meat products. Although the microbiological contaminating organisms prevalent in commercial abattoirs for the traditionally farmed species have been documented, very little data exist on those found in commercial ostrich abattoirs2.
This study focuses on ostrich meat and as reported6,7, the main export product of the ostrich industry is fresh meat and it is therefore important to have as long a shelf-life as possible. One effective way to ensure a good shelf-life is to ensure a low initial microbial load12. Karama10 reported that there was no significant increase in microbial count on carcasses from postflaying to post-evisceration, but a significant increase was detected from post-evisceration to post-chilling. Bearing this in mind, prevalent organisms were isolated from bruised areas on the carcasses directly post-evisceration and from hot-trimmed exposed meat after 24 hours chilling in the de-boning area. This was done to get an indication of microbial populations found on the ostrich carcasses at different processing points. To monitor for possible environmental contamination, samples from work surfaces, different water sources and personnel were taken in the same production areas.
Selected microorganisms from the meat samples as well as the environmental samples were identified. It was attempted to establish the major cause of contamination and in this way equip abattoir management with a strategy to prevent meat contamination and increase shelflife.
MATERIALS AND METHODS
Abattoir
In a modern European Union exportapproved ostrich abattoir in South Africa (Klein Karoo International Abattoir 1, ZA92), commercially reared ostriches were slaughtered, their feathers plucked, the skins removed (flaying) and the intestines removed (eviscerating)8. In this abattoir, the thighs are removed from the carcasses post-evisceration and the ribcages are discarded (hot de-boning), as only meat from the thighs is utilised commercially. The thighs are moved into an overnight chiller operating at 0-4 ºC, where they are cooled for approximately 24 hours.
Trials and sampling
This study comprised 5 trials, each performed over a 2-day period. The bacteria from only 2 of the 5 trials, 1 performed in summer and the other during late autumn, were characterised and identified. Suitable experimental carcasses with bruises on the thigh or back muscles were selected at the primary meat inspection point, just after the evisceration process. At least 6 carcasses were identified. Bruises were either trimmed on hot carcasses (at the primary meat inspection point) or on chilled carcasses (left to chill overnight and then trimmed at the de-boning department). Table 1 gives an overview of the sampling procedure.
Environmental samples were taken along the entire slaughter line. The air samples in each area were taken by placing open agar plates in the area for 10 min. The water from taps, hoses and water brooms were also collected in sterile specimen jars, while water from the drains and platforms in the evisceration and primary meat inspection areas were collected in sterile syringes. The swabs on work surfaces, workers' hands and knives were taken by the Rodac plate method11.
Handling of samples
The meat samples were placed in marked sterile stomacher bags and stored on ice along with the specimen jars and Rodac plates. All the samples were transported in the coolers to the Klein Karoo International Research Laboratory, where they were analysed on the same day in accordance with South African National Standards (SANS); these standards are based on the international ISO standards20,21.
Microbial analysis
At the laboratory a dilution series was prepared for each sample, all the samples were plated out or drawn at the abattoir (Rodac plates) on 3 selected media: Plate Count Agar (PCA, Biolab code C6), Violet Red Bile Glucose Agar (VRBG Oxoid code CM485) and MRS (MRS, Biolab code C86). The PCA plates were incubated for 48hoursat30± 1ºC,theVRBGat37± 1 ºC for 24 hours and the MRS at 37±1ºC for 48 hours. On the PCA plates the aerobic viable counts were evaluated to provide an indication of both Gram-positive and negative organisms19. From the VRBG plates the Enterobacteriaceae counts22 were obtained and only Gramnegative rods were expected to grow. The MRS medium was not acidified or incubated under anaerobic or micro-aerophilic conditions as it was decided to rather use the medium's enrichment properties than its selective properties for lactobacilli. The expected growth on these plates included Lactobacillus, Streptococcus, Pediococcus and and Leuconostoc, which are all Gram-positive catalase and oxidase negative organisms.
Isolation of colonies
Colonies from the above plates were selected for identification. On plates with fewer than 5 colonies, all colonies that were morphologically unique were isolated. On plates with heavier growth, the Harrison's disc method5 was used to select colonies. In this procedure the plates were superimposed on the disc proposed by Harrison and the colonies that fell inside the demarcated vectors were typed. With this method it is possible to calculate the percentage distribution of various organisms on the plates when it is not possible to type all of the organisms. Selected colonies were aseptically picked off the plates by making use of a sterile inoculation loop.
All selected colonies were plated out for single colonies on blood agar plates (Columbia agar, Oxoid code CM331 with addition of 5 % ostrich blood). These plates were incubated at 37 ± 2 ºC for 48 h.
Characterisation
Isolated pure colonies were removed from these plates for Gram staining according to Preston and Morrel's15 method. During microscopic inspection of the stained slides, Gram-positive organisms, Gram-negative organisms and yeasts were distinguished. The bacterial morphology was also recorded, i.e. cocci, rods, etc. The Gram-positive colonies were tested for catalase and the Gram-negatives were tested for oxidase activity.
Identification
On the basis of the above information, the colonies were identified using the Remel RapID systems17. Each of the RapID systems has vials with either 1 or 2m of inoculation fluid. The fluids were adjusted by the addition of pure colonies to a density specified for each system, based on the McFarland Equivalence Turbidity Standards. The turbid inoculation fluid was transferred with a glass pipette into the appropriate RapID identification panel with dehydrated reagents for biochemical identification reactions in the wells. The panels with the wells were tilted, 1st to the side to distribute the fluid evenly and eliminate possible air bubbles and then forward to fill up the wells. The reagents dissolved and during incubation reacted with the organisms present in the fluid to produce easy to interpret colour reactions from the degradation of both chromogenic and conventional substrates. The panels were incu-bated according to prescribed procedures, in most instances for 4 hours at 37 ± 2 ºC16. After incubation the RapID Colour guides were used to read and score each well on the grounds of a positive or negative reaction. The scores for the panels were electronically captured into the ERIC (Electronic RapID Compendium)18, developed to process micro codes for all the RapID identification systems16. ERIC processed the micro codes and gave the microorganism options, including probability percentages, on the grounds of the reactions in the wells.
Out of the 8 individual RapID systems, the following 6 systems were used: RapID ONE for the identification of Enterobacteriaceae and other oxidase negative bacteria; RapID CB Plus for the identification of Corynebacterium, Actinomyces and other related bacteria; RapID NF Plus for the identification of glucose fermenting and non-fermenting Gram-negative bacteria not belonging to the family Enterobacteriaceae; RapID Yeast Plus for the identification of medically important yeasts; RapID STR for the identification of streptococci and other similar Gram-positive bacteria. All Gram-positive, catalase positive coccoid morphological colonies were tested on the Oxoid Staphylases test kit (Oxoid code DR595A) to identify Staphylococcus aureus colonies13.
RESULTS
Enumeration of colonies
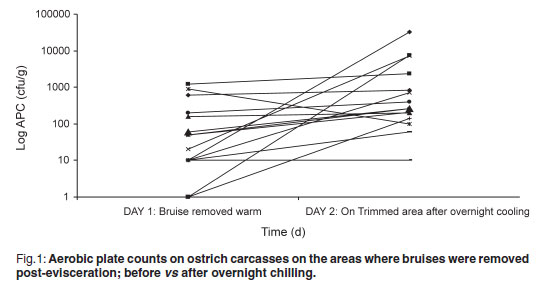
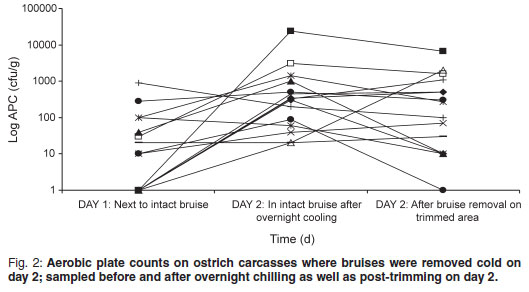
The aerobic plate count (APC) found on carcasses on the slaughter-line was 219.47 cfu/g on carcasses where the bruises were already trimmed and 99.73 cfu/g on carcasses where the bruises were left intact. On the warm-trimmed carcasses, counts increased from 219.47 cfu/g to an average level on day 2 of 3494.07 cfu/g (Fig. 1); the cold-trimmed carcasses surface values increased from 99.73 cfu/g to 2142.00 cfu/g on day 2, after overnight cooling, and then decreased to 887.93 cfu/g after cold trimming (Fig. 2).
Identification
A total of 198 organisms were selected on the basis of Gram stain. These organisms were collected from both carcass and environmental samples. Of these, 91 organisms were found to be Grampositive, 82 Gram-negative and 25 were identified as yeasts. Only 1 mould was found. Out of the 198 organisms that were Gram stained, 161 were further evaluated for oxidase or catalase activity to determine on which RapID systems they should be inoculated. Of the 91 Grampositive organisms, 74 were catalase positive and 9 were catalase negative. Of the 82 Gram-negative bacteria, 25 were found to be oxidase positive and 53 oxidase negative.
For the identification of the microorganisms, the organisms isolated from carcasses were divided into 2 groups, post-evisceration (day 1) and post-chilling (day 2). The prevalent growth on the bruised carcasses directly post-evisceration was predominantly Gram-positive organisms (Table 2).
After overnight cooling to 4 ºC, the bacterial counts were on average 10 times higher than on the post-evisceration samples and more isolates were selected for identification which indicated a pronounced presence of Gram-negative bacilli. Out of the 41 organisms isolated from carcasses post chilling, 9 were identified as yeasts (Table 2). Many of the yeasts could not be effectively identified because the RapID Yeast Plus systems were only obtained later during the study and it only covers options for yeasts associated with human medicine.
The isolates from the air sample plates had both Gram-positive (15) and Gramnegative (20) organisms. On the contact plates of the 1st trial (summer), the growth was predominantly Gram-negative (66.67 % of isolates), while on that of the 2nd trial (autumn), apart from the Enterobacteriaceae (VRBG?) plates, the growth was markedly more Gram-positive (63.89 % of isolates from PCA and MRS plates). Also, in the 2nd trial a very high number of yeast colonies (69.23 % of isolates, n = 9) were isolated from the MRS media. If the colonies on the contact plates are split into contamination from workers and that from surfaces for both trials, the prevalent growth is evenly distributed between Gram-positive and negatives, with a significant yeast population as well. The growth on water samples tended to be predominantly Gram-negative (81.82 % of isolates, n = 11).
The prevalent organisms isolated from ostrich carcasses post evisceration were the Gram-positive cocci (64.29 % of isolates, n = 14), Gemella morbillorum (4 isolates) closely resembling the Streptococcus spp. and Pediococcus pentosaceus (3 isolates), a member of the lactic acid bacteria group23. The samples of carcasses post-chilling also had a wide variety of Gram-negative organisms.
The Gram-positive isolates from the air sample plates were primarily Gemella morbillorum.The Gram-negative organisms on the air plates consisted primarily of Shigella spp. Proteus sp. was also isolated from the air plates and the air supply in the abattoir. Gemella morbillorum was isolated from the cold rooms and deboning area.
The prevalent Gram-positive organisms isolated from both surfaces and workers were Gemella (11 out of 44) and Pediococcus (13 out of 44). The Gram-negative organisms consisted of predominantly Shigella and Serratia spp., but also included Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas and other organisms.
From the successfully identified yeast species, more than 90 % were typed as a Cryptococcus sp., a type of yeast that is known to occur on fresh refrigerated meat9.
In this abattoir, the water from taps, hoses and water brooms are all from the water reticulation system that is treated with chlorine dioxide and no organisms were found in these water samples. The water collected from drains and platforms showed very high counts (>2 × 103 cfu/mℓ on all 3 media) and the growth was predominantly Gram-negative (81.82 % of isolates, n = 11) (Table 2).
DISCUSSION
The APC found on the carcasses are very low when compared with those found by Harris et al.4 and Karama10 (APC values averaging 3.2 to 4.21 log cfu/cm2 on groups of carcasses post-evisceration). All 3 post-chill average APC values were still significantly lower than the 4.57 log cfu/cm2 reported by Karama10 for post-chilling samples. From Figs 1 and 2 it is evident from the decrease in microorganisms after the cold trim that it is beneficial from a microbiological point of view to cold trim rather than warm trim carcasses. Karama10 was therefore correct in stating that the biggest increase in bacterial growth is from post-evisceration to post-chilling (Fig. 1).
The presence of Gram-negative bacilli on the chilled samples indicates that the contaminants were both Gram-positive and Gram-negative, but that some or all of the Gram-negative organisms were probably more psychrophilic and grew better under refrigerated conditions. The Gram-negative Shigella spp. found on the air plates are most probably from the evisceration hall, because these organisms are associated with the gastrointestinal tract of animals and not with the environment9. The Proteus sp. that was also isolated from the air plates and the air supply in the abattoir could possibly contribute to the Gram-negative contamination of the carcasses. The Stenotrophomonas maltophilia isolated from the cold rooms and de-boning areas are associated with the pseudomonads14 and are thus able to grow and contaminate the meat in these temperature controlled areas.
Many of the organisms found in the water from drains and platforms were human pathogens (Shigella, E. coli and Salmonella) and even though E. coli and Salmonella were not recovered from the carcasses in these trials, the standing water in the drains and on the platforms pose a very real risk to food safety and should be prevented as far as possible.
All of the yeasts isolated, were isolated during the trial that was conducted late in the production season (autumn) and all 8 were collected from samples in the de-boning hall on day 2 after chilling. Jay9 indicated that yeasts can grow over a wide range of temperatures and relative humidity, thus, chiller conditions would also support the growth of yeasts. Yeast colonies were also isolated from almost all the environmental samples, with a high incidence on the surface swabs. More yeast isolates were found post-chilling which indicated that the carcasses were contaminated with these organisms, most probably from the knives and workers.
According to Jay9, all the above-mentioned microorganisms have the environment, animals and/or humans as important sources. These results confirm what Harris et al.4 reported regarding the growth on ostrich carcasses, i.e. it is mostly associated with organisms native to the environment and the skin of animals and humans. No Salmonella colonies were isolated from the carcasses, which corresponds with a previous study1 that found no Salmonella on any ostrich meat intended for human consumption.
Some of the organisms selected died off during isolation or there were not enough options under the Remel RapID systems to facilitate their successful identification. As the Remel RapID systems were developed specifically for human clinical specimens they focus only on microorganisms associated with human diseases and infections. During the use of these systems for the identification of organisms in the abattoir and on the meat it was found that while there were enough options on the Gram-negative side to ensure the successful typing of all organisms, there were limited options to identify the Gram-positive organisms.
CONCLUSIONS
Based on the findings of this study, it can be recommended that proper environmental management in the abattoir can be used to control the microbiological load on ostrich meat before packaging to a large extent. Removing bruised meat cold during de-boning is recommended as well as controlling the external sources of microbiological contamination inside the slaughter-house. With regard to the management of contamination, a programme for water containment is of major importance. This should include the prevention of pooling of water or blockage of drains and the control over the usage of high-pressure hoses; subsequently a programme for the general hygiene of the abattoir cannot be neglected. This should focus on good manufacturing practices and include the implementation of filtered air supply and regular cleaning of air ducting, the cleaning and sanitising of work surfaces and equipment and personnel hygiene.
REFERENCES
1. Gopo J M, Banda G N 1997 Occurrence of Salmonella on meat and products in an ostrich abattoir as determined with a DNA probe. South African Journal of Animal Science 27: 1-6 [ Links ]
2. Gill C O 2007 Microbiological conditions of meats from large game animals and birds. Meat Science 77: 149-160 [ Links ]
3. Grau F H 1986 Microbial ecology of meat and poultry. In Pearson A M, Dutson T R (eds) Advances in meat research Vol. 2. Michigan State University, East Lansing, MI: 1-36 [ Links ]
4. Harris S D, Morris C A, Jackson T C, May S G, Lucia L M, Hale S D, Miller R K, Keeton J T, Savell J W, Acuff G R 1993 Ostrich meat industry development, Final report to: American Ostrich Association. Texas Agricultural Extension Service 348, Kleberg: 14 [ Links ]
5. Harrigan W F, McCance M E 1976 Preface In Laboratory methods in food and dairy microbiology (rev. edn.). Academic Press, London: 47-49 [ Links ]
6. Hoffman L C 2005 A review of the research conducted on ostrich meat. In: Proceedings of the 3rd International Ratite Science Symposium of the WPSA, Madrid, Spain, October 2005: 107-119 [ Links ]
7. Hoffman L C 2008 Value adding and processing of ratite meat: a review. Australian Journal of Experimental Agriculture 48: 1-6 [ Links ]
8. Hoffman L C, Fisher P 2001 Comparison of meat quality characteristics between young and old ostriches. Meat Science 59: 335-337 [ Links ]
9. Jay J 1992 Modern food microbiology (4th edn) Chapman and Hall, New York [ Links ]
10. Karama M 2001 The microbial quality of ostrich carcasses produced at an exportapproved South African Abattoir. MMed-Vet (Hyg) thesis, University of Pretoria, Pretoria, South Africa [ Links ]
11. Lemmen S W, Hafner H, Zolldann D, Amedick G, Lutticken R 2001 Comparison of 2 sampling methods for the detection of gram-positive and gram-negative bacteria in the environment: moistened swabs versus Rodac plates. International Journal of Hygiene and Environmental Health 20: 245- 248 [ Links ]
12. McKinnon D C, Heyneke P G, Olivier A J, Mulder C, Britz T J, Hoffman L C 2005 Effect of ozone and UV on the microbial load of muscle surfaces of ostrich carcasses. Proceedings of the 3rd International Ratite Science Symposium of the WPSA, Madrid, Spain, October 2005: 276 [ Links ]
13. Oxoid 2003 Diagnostic reagents: staphylase test. Oxoid, Basinstoke, Hampshire, England [ Links ]
14. Pearson A 2005 Pseudomonas spp. and Stenotrophomonas maltophilia bacteraemia: England, Wales, and Northern Ireland: 2004. CDR Weekly 15(39): 1-6 [ Links ]
15. Preston N W, Morrell A 1962 Reproducible results with the Gram stain. Journal of Pathology and Bacteriology 84: 241-243 [ Links ]
16. Remel 2000 IDS RapID Systems. Lenexa, KS, USA [ Links ]
17. Remel 2005 RapID Systems. Lenexa, KS, USA [ Links ]
18. Remel 2005 RapID with ERIC means results. Lenexa, KS, USA [ Links ]
19. SANS ISO 15214 1998 Horizontal method for the enumeration of mesophilic lactic acid bacteria - Colony count technique at 30ºC. South African Bureau of Standards, Pretoria [ Links ]
20. SANS ISO 6887-1 1999 Preparation of test samples, initial suspension and decimal dilutions for microbiological examination - general rules. South African Bureau of Standards, Pretoria [ Links ]
21. SANS 11133 2004 Microbiology of food and feeding stuffs - Guidelines on preparation and production of culture media. South African Bureau of Standards, Pretoria [ Links ]
22. SANS 218528 2005 Microbiology of food and feeding stuffs - Horizontal method for the detection and enumeration of Enterobacteriaceae. South African Bureau of Standards, Pretoria [ Links ]
23. SneathPHA, Mair N S, Sharpe M E, Holt J G 1986 Bergey's manual of systematic bacteriology, Vol. 2. Williams and Wilkens, Baltimore, USA [ Links ]
Received: July 2010.
Accepted: July 2010.
* Author correspondence. E-mail: lch@sun.ac.za














