Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
Journal of the South African Veterinary Association
versión On-line ISSN 2224-9435
versión impresa ISSN 1019-9128
J. S. Afr. Vet. Assoc. vol.81 no.2 Pretoria ene. 2010
ARTICLE ARTIKEL
The prevalence of ovine herpesvirus-2 in 4 sheep breeds from different regions in South Africa
C W Bremer
Biotechnology Division, ARC-Onderstepoort Veterinary Institute, Private Bag X05, Onderstepoort, 0110 South Africa. E-mail: vroonw@arc.agric.za
ABSTRACT
About 90 % of bovine malignant catarrhal fever (BMCF) PCR-positive cases in South Africa are caused by alcelaphine herpesvirus-1 (AlHV-1) and the other 10 % by ovine herpesvirus-2 (OvHV-2). The prevalence of OvHV-2 in different sheep breeds in South Africa was determined in order to investigate whether the lower incidence of BMCF caused by OvHV-2 in comparison with AlHV-1 can be ascribed to a low incidence of the virus in sheep. A single-tube hemi-nested PCR was developed, evaluated and applied to detect OvHV-2 DNA. The prevalence of the virus in 4 sheep breeds from various regions in South Africa was shown to be 77 %. No statistically significant difference was found amongst the sheep breeds tested.
Keywords: bovine malignant catarrhal fever, ovine herpesvirus-2, PCR, prevalence, sheep.
INTRODUCTION
Malignant catarrhal fever (MCF) is a lymphoproliferative disease of cattle, pigs and certain species of captive and wild ruminants and has worldwide distribution. The causative agents are ruminant rhadinoviruses of the Gammaherpesvirinae subfamily, genus Rhadinovirus5. Alcelaphine herpesvirus-1 (AlHV-1) is carried asymptomatically by wildebeest (Connochaetes taurinus and C. gnou)15,17 and ovine herpesvirus-2 (OvHV-2) by sheep4. Both AlHV-1 and OvHV-2 cause disease in cattle. OvHV-2 also causes disease in various other ruminant species as well as in pigs12.
In the USA the prevalence of OvHV-2 DNA in adult sheep was 99 % as determined by nested polymerase chain reaction (PCR)10. In South Africa, approximately 90 % of MCF PCR-positive cases are associated with AlHV-1 and only 10 % with OvHV-2 (M. Romito, Agricultural Research Council-Onderstepoort Veterinary Institute (ARC-OVI) pers. comm. 2009). As a result, most of the studies on MCF in cattle in South Africa have been done on AlHV-1. One study indicated the presence of AlHV-1 in 100 % of wildebeest calves by using a nucleic acid probe13 while in another study AlHV-1 was isolated from 65 % of wildebeest calves1.
A better understanding of the epidemiology of OvHV-2 in South Africa is required. Although the prevalence of OvHV-2 DNA in sheep in the USA was shown to be 99 %, this may not be true for sheep in South Africa. The purpose of this investigation was to determine whether OvHV-2 DNA could be detected in 4 different sheep breeds in South Africa, and if positive, to determine the prevalence of OvHV-2 DNA.
MATERIALS AND METHODS
Blood and DNA samples
Sheep. Heparinized blood samples (n = 85) were collected from female adult sheep from various regions in South Africa.
Cattle. DNA extracted from positive bovine blood (n = 12) samples used for validation of the hemi-nested PCR was obtained from Dr Marco Romito of the ARC-OVI. The blood samples were sent to the Institute to confirm the presence of OvHV-2 DNA by PCR using a method described previously7.
Cell cultures
Bovine herpesvirus-1 (BHV-1) and bovine herpesvirus-2 (BHV-2) were cultivated in VERO cells and bovine herpesvirus-4 was cultivated in foetal bovine testes cells. Cells were cultivated in Eagles Minimum Essential Medium supplemented with 2 mM L-glutamine, 0.1 mM non-essential amino acids, ~100 U/mℓ penicillium, ~100 µg/mℓ streptomycin and 5-7 % foetal calf serum (Gibco).
Virus strains
BHV-1, -2 and -4 were field isolates from cattle that were cultivated in cell culture and identified by PCR. With the exception of the primers, the PCR reaction mixes contained the same reagents as the OvHV-2 hemi-nested PCR. The PCR primers used have been described previously6,8,18. Reaction conditions for PCRs were BHV-1: 94 ºC 2 min (1×) followed by 94 ºC/30 s, 54 ºC/45 s, 72 ºC/45 s (45×), BHV-2: 94 ºC 2 min (1×) followed by 94 ºC/30 s, 50 ºC/30 s, 72 ºC/30 s (45×), BHV-4: 94 ºC 2 min (1×) followed by 94 ºC/30 s, 54 ºC/30 s, 72 ºC/30 s (45×).
DNA extraction from sheep and bovine samples
Buffy coats of sheep blood samples in heparin were prepared by centrifugation of 1mℓ blood at 540× g for 20 min. The top layer of each blood sample (approximately 0.2 mℓ ) was removed and placed in an Eppendorf tube. DNA was extracted from this using the NucleonTM BACC genomic DNA extraction kit (Amersham Biosciences) according to the manufacturer's protocol. Bovine blood samples used as control samples were either whole blood (collected in EDTA or heparin tubes) or blood clots (collected in tubes without additive). DNA was extracted using the NucleonTM BACC genomic DNA extraction kit (Amersham Biosciences) or the MagNa Pure LC total nucleic acid isolation kit (MagNa Pure LC instrument, Roche).
Single-tube hemi-nested PCR procedure
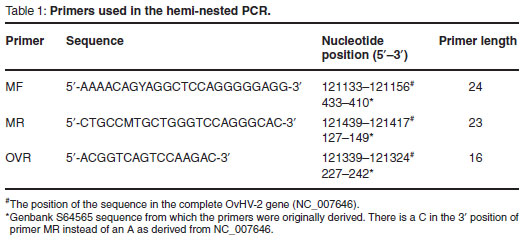
For the detection of OvHV-2 DNA a single-tube hemi-nested PCR format was used which was based on the single-tube duplex nested PCR described previously3, in which the primers targeted the ORF 75 gene. In the OvHV-2 specific single-tube hemi-nested PCR only primers MF, MR and OVR were used. In both steps of the single-tube hemi-nested PCR, samples were subjected to additional amplification steps with the aim of further increasing the sensitivity of the procedure.
PCR reactions were performed in a volume of 12.5 µℓ consisting of 1 x ExTaq DNA polymerase buffer, 0.5 U ExTaq DNA polymerase, 200 µM of each dATP, dCTP, dGTP and dTTP (buffer, enzyme and dNTPs were from TaKaRa Bio Inc, Japan), 0.08 µM primer MR and 0.8 µM of primers MF and OVR respectively and approximately 50-150 ng total DNA. The reaction conditions were 25 cycles of 94 ºC/30 s, 68 ºC/30 s, 72 º C/30 s followed by 45 cycles at 94 ºC/30 s, 54 ºC/30 s, 72 ºC/30 s. PCR products were analysed by electrophoresis in 1.5 % agarose gels in 40 mM Trisacetate, 1 mM EDTA buffer.
Restriction enzyme digestion
Approximately 1/3rd of the amplified DNA product of each sample was digested for 3-24 h at 37 ºC with approximately 0.3 U Alu I enzyme in the appropriate digestion buffer in a total volume of 20 µℓ .
Samples were analysed by electrophoresis in a 12 % polyacrylamide gel9, which was rinsed in water for 5-10 min and stained in approximately 10 ng/µℓ ethidium bromide in water or in Laemmli buffer without SDS.
Construction of a recombinant pGEM-T vector containing OvHV-2 DNA
With the purpose of determining the sensitivity of the hemi-nested PCR, a recombinant pGEM-T vector was prepared. PCR was performed on DNA extracted from a positive OvHV-2 field sample using outer primers MF and MR only. The PCR product was subjected to agarose gel electrophoresis, excised from the gel and purified using the QIAEX gel purification kit (Qiagen, Hilden, Germany). The product was then inserted directly into the pGEM-T vector since TaKaRa Ex Taq enzyme used for the PCR adds a single dATP onto the 3' end. After transformation, the recombinant vector was cloned and purified and dilutions were made for the determination of sensitivity.
Statistics
In this study, the presence of viral DNA in different sheep breeds was tested and classified as positive or negative. To determine whether the distribution of positive to negative is the same for each breed and vice versa, i.e. to determine whether the 2 types of attributes are independent, the Chi-square test was performed.
RESULTS
Evaluation of the hemi-nested PCR
Specificity
The specificity of the PCR was determined by performing PCR on DNA extracted from monolayer mammalian cells infected with BHV -1, -2 and -4, from a blood sample of a bovine infected with AlHV-1, as well as from an uninfected animal that tested negative using PCR. No bands of the expected sizes were observed during analysis of the PCR products of the related BHV samples (results not shown). A band of 307 bp was occasionally seen with AlHV-1 samples as a result of the conserved regions in the genes of the 2 viruses on which primers MF and MR were based3. The absence of the band in some samples was probably as a result of a low concentration of AlHV-1 DNA in the samples.
For further confirmation of the specificity of the PCR, amplicons of each positive sample were subjected to restriction enzyme digestion making use of an Alu I restriction site present at position 102 in the sequence of the 207 bp nested PCR amplicon, yielding a 102 bp and a 105 bp fragment which could not be distinguished using SDS-PAGE based on the method of Laemmli 19709. In Fig. 1 undigested (lanes 1 and 3) and digested (lanes 2 and 4) PCR amplicons of the expected sizes can be observed. In the sample that also contained the 307 bp PCR product (lane 3), 2 bands were observed after digestion: the 1st consisting of the 102 and 105 bp fragments, and the 2nd of a 205 bp fragment (lane 4).
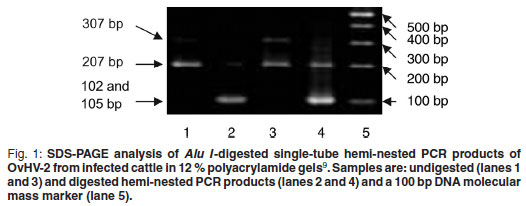
Sensitivity
The sensitivity of the single-tube heminested PCR was determined by performing the assay on 10-fold dilutions of DNA isolated from a recombinant pGEM-T clone. Dilutions were made of the purified recombinant pGEM-T plasmid containing the OvHV-2 DNA specific sequence and a 10 µℓ aliquot of each dilution was added to 90 µℓ of blood. A total DNA extraction was done on the spiked blood. The detection limit of the recombinant plasmid pGEM-T in bovine blood was 0.5 fg (result not shown).
Validation
To validate the assay, single-tube heminested PCRs were performed on 12 DNA samples isolated from the blood of OvHV-2 PCR-positive bovine field samples using the method of Dungu et al.7. These were all shown to be positive after analysis of PCR amplicons by agarose gel electrophoresis (results not shown).
Detection of OvHV-2 DNA in sheep
The single-tube hemi-nested PCR was performed on DNA isolated from blood samples (n = 85) of 4 different sheep breeds as described above. Results of samples from Döhne Merino are shown in Fig. 2. A band of the expected size (207 bp) produced by hemi-nested PCR was observed in observed in 7 of the 8 sheep samples shown (lanes 1–8) as well as in the positive control sample (lane 9). Two extra bands, 1 of 307 bp, the expected size of the 1st PCR, as well as an approximately 600 bp band, were also observed in the positive control sample (lane 9). The band of approximately 600 bp is probably non-specific, because of the high number of amplification cycles. Samples producinging only the 207 bp band probably contained a low concentration of target DNA as PCR products obtained from the lowest concentrations of DNA in the sensitivity determination contained a single 207 bp band only but samples with a higher concentration of DNA contained extra bands (results not shown).
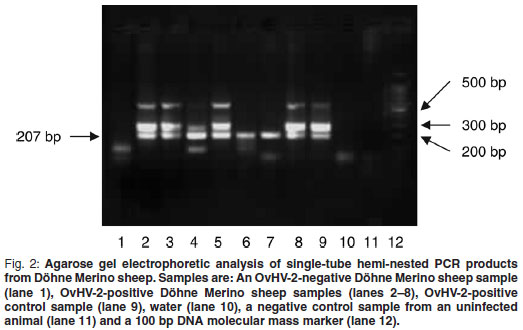
All samples produced were subjected to Alu I restriction enzyme digestion (Fig. 1) and the expected products were obtained.
Table 2 shows results obtained when single-tube hemi-nested PCR was performed on DNA extracted from the blood of different sheep breeds.

Statistical analysis
Statistical analysis of the data in Table 1 using the Chi-square test indicated that differences were not significant (χ2= 2.038; P = 0.568; d.f. = 3). The Dorper sheep, however, showed the largest positive response (85.7 %) and Karakul sheep the smallest positive response (63.6 %).
DISCUSSION
As a result of the problem of contamination associated with a 2-step nested PCR, a single-tube hemi-nested PCR was developed for the detection of OvHV-2 DNA. The method was validated and shown to be suitable for the detection of OvHV-2 in field samples.
The sensitivity of the test was in the range obtained by other laboratories and as little as 0.5 fg of the recombinant plasmid pGEM-T could be detected in bovine blood. A nested PCR used by Li et al.10 could detect 10 fg of pUC18 containing the 238 bp fragment after analysis of the product by agarose gel electrophoresis and after Southern blot hybridization 0.4 fg could be detected.
One study found that 99 % of adult sheep from the USA were positive for OvHV-210 when using the 2-step heminested PCR described previously2.In India, OvHV-2 DNA was detected in 84.8 % of adult sheep, using hemi-nested PCR19. In South Africa, where the detection rate in sheep was 77 %, cattle and sheep are not often stabled together and the chances of transmission of the virus are probably smaller than in the USA where cold conditions may require that cattle and sheep be stabled in the same area. Furthermore, the South African OvHV-2 strains and their infectivity for South African sheep breeds may differ from those of USA strains for USA sheep breeds. Despite the high average infection rate in the different sheep breeds, OvHV2 DNA is detected in only about 10 % of PCR positive field cases of clinical MCF in cattle in South Africa (M. Romito, ARC-OVI, pers. comm. 2009). In one study individual adult sheep were shown to have relatively elevated levels of OvHV-2 DNA in nasal secretions and blood and this suggested that adult sheep might be intermittent high shedders of the virus11. The sporadic nature and low incidence of the disease in cattle caused by OvHV-2 are thought to be related to the varying levels of viral DNA in nasal secretions in adult sheep . Climatic factors that affect the survival of the virus in the environment, presence of vectors, variation in stocking density and levels of stress due to weather or management practices, have also been suggested as factors that may influence the incidence rate.
Cattle seem to be more resistant to the development of clinical signs after infection with OvHV-2 than with AlHV-1. The association between OvHV-2 infection and the development of clinical MCF has been studied in cattle housed at a dairy where bovine MCF had been previously diagnosed and where they were kept in close proximity to sheep16. None of the cattle showed clinical signs, but 70 % of the cattle had evidence of infection as determined by competitive inhibition enzyme-linked immunosorbent assay or PCR during a 20-month surveillance period. In a similar experiment, 3 cattle that were housed with a viraemic wildebeest died of MCF14. However, when 2 other cattle were housed with the same wildebeest after shedding of the virus had stopped, they did not develop clinical MCF.
This is the 1st study in which the prevalence of OvHV-2 in different sheep breeds in South Africa has been determined. OvHV-2 was detected in all 4 sheep breeds tested and no significant difference in the infection rate of different breeds was indicated.
ACKNOWLEDGEMENTS
The author thanks Drs M. Romito and G.H. Gerdes for providing the samples and reviewing the manuscript, Dr A.C. Potgieter for the preparation of the pGEMT clone, Ms M. Smith for statistical analyses and Prof. G.J. Viljoen for valuable discussions. This work was funded by the Red Meat Research and Development Trust (RMRDT).
REFERENCES
1. Barnard B J H, Van de Pypekamp H E, Griessel M D 1989 Epizootiology of wildebeest-derived malignant catarrhal fever in an outbreak in the north-western Transvaal: indications of an intermediate host. Onderstepoort Journal of Veterinary Research 56: 135-139 [ Links ]
2. Baxter S I F, Pow I, Bridgen A, Reid H W 1993 PCR detection of the sheep-associated agent of malignant catarrhal fever. Archives of Virology 132: 145-159 [ Links ]
3. Bremer C W, Swart H, Doboro F A, Dungu B, Romito M, Viljoen G J 2005 Discrimination between sheep-associated and wildebeest-associated malignant catarrhal fever virus by means of a single-tube duplex nested PCR. Onderstepoort Journal of Veterinary Research 72: 285-291 [ Links ]
4. Bridgen A, Reid H W 1991 Derivation of a cDNA clone corresponding to the viral agent of sheep-associated malignant catarrhal fever. Research in Veterinary Science 50: 38-44 [ Links ]
5. Crawford T B, Li H, Rosenburg SR, Norhausen R W, Garner M M 2002 Mural folliculitis and alopecia caused by infection with goat-associated malignant catarrhal fever virus in two sika deer. Journal of the American Veterinary Medical Association 221: 843-847 [ Links ]
6. D'Offay J M, Floyd J G Jr, Eberle R, Saliki J T, Brock K V, D'Andrea G H, McMillan K L 2003 Use of a polymerase chain reaction assay to detect bovine herpesvirus type 2 DNA in skin lesions from cattle suspected to have pseudo-lumpy skin disease. Journal of the American Veterinary Medical Association 222: 1404-1407 [ Links ]
7. Dungu B, Bosman A-M, Kachelhoffer C, Viljoen G J 2002 Single-tube nested PCR for detection of the sheep-associated agent of MCF. Veterinary Record 151: 703-706 [ Links ]
8. Egyed L, Ballagi-Pordany A, Bartha A, Belák S 1996 Studies of in vivo distribution of bovine herpesvirus type 4 in the natural host. Journal of Clinical Microbiology 34: 1091-1095 [ Links ]
9. Laemmli U K 1970 Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [ Links ]
10. Li H, Shen D T, O'Toole D, Knowles D P, Gorham J R, Crawford T B 1995 Investigation of sheep-associated malignant catarrhal fever virus infection in ruminants by PCR and competitive inhibition enzyme-linked immunosorbent assay. Journal of Clinical Microbiology 33: 2048-2053 [ Links ]
11. Li H, Hua Y, Snowder G, Crawford T B 2001 Levels of ovine herpesvirus 2 DNA in nasal secretions and blood of sheep: implications for transmission. Veterinary Microbiology 79: 301-310 [ Links ]
12. Loken T, Alexandersen M, Reid H, Pow I 1998 Malignant catarrhal fever caused by ovine herpesvirus-2 in pigs in Norway. Veterinary Record, 143: 464-467 [ Links ]
13. Michel A L 1993 Generation of a nucleic acid probe specific for the alcelaphine herpesvirus 1 and its use for the detection of malignant catarrhal fever virus DNA in blue wildebeest calves (Connochaetes taurinus). Onderstepoort Journal of Veterinary Research 60: 87-93 [ Links ]
14. Plowright W 1965 Malignant catarrhal fever in East Africa. II. Observations on wildebeest calves at the laboratory and contact transmission of the infection to cattle. Research in Veterinary Science 6: 69-83 [ Links ]
15. Plowright W, Ferris R D, Scott G R, 1960 Blue wildebeest and the aetiological agent of bovine malignant catarrhal fever. Nature 188: 1167-1169 [ Links ]
16. Powers J G, Van Metre D C, Collins J K, Dinsmore R P, Carman J, Patterson G, Brahmbhatt D, Callan R J 2005 Evaluation of ovine herpesvirus type 2 infections, as detected by competitive inhibition ELISA and polymerase chain reaction assay, in dairy cattle without clinical signs of malignant catarrhal fever. Journal of the American Veterinary Medical Association 15: 606-611 [ Links ]
17. Pretorius J A, Oosthuizen M C, Van Vuuren M 2008. Gammaherpesvirus carrier status of black wildebeest (Connochaetes gnou)in South Africa. Journal of the South African Veterinary Association 79: 136-141 [ Links ]
18. Vilcek S 1993 Detection of the bovine herpesvirus-1 (BHV-1) genome by PCR. Journal of Virological Methods 41: 245-247 [ Links ]
19. Wani S A, Samanta I, Pandit F, Buchoo B A, Peer F, Bhat M A 2006 Molecular epidemiology of ovine herpesvirus type 2 infection in Kashmir, India. The Veterinary Record 159: 587-590 [ Links ]
Received: January 2010.
Accepted: May 2010.














