Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Chemistry
versión On-line ISSN 1996-840X
versión impresa ISSN 0379-4350
S.Afr.j.chem. (Online) vol.77 Durban 2023
http://dx.doi.org/10.17159/0379-4350/2023/v77a08
RESEARCH ARTICLE
Mononuclear discrete Ag(I) complexes of aryl substituted (E)-N-phenyl-1-(pyridin-3-yl)methanimine: In vitro biological activities and interactions with biomolecules
Adesola A. AdelekeI, II; Md. Shahidul IslamIII; Kolawole OlofinsanIII; Veronica F. SalauIII; Bernard OmondiI,*
ISchool of Chemistry and Physics, University of KwaZulu-Natal, South Africa
IIDepartment of Chemical Sciences, Olabisi OnabanJo University, Ago-Iwoye, Nigeria
IIIDiscipline of Biochemistry, School of Life Sciences, University of KwaZulu-Natal, Durban, South Africa
ABSTRACT
The synthesis of three pyridinyl imines (L1-L3) with electron-donating and electron-withdrawing functional groups, as well as their silver(I) complexes (1a-3a, 1b-3b and 1c-3c), resulted from our ongoing search for novel metallodrugs with potential pharmacological activity. Various analytical and spectroscopic analyses were used to characterize the Ag(I) complexes. The single crystal X-ray diffraction analysis of complexes 3a and 3b in the solid state confirmed their linear geometry around the Ag(I) center. In vitro antibacterial properties of the complexes evaluated using the minimum inhibitory concentration (MIC) method revealed that complexes containing either a Me- or an F-substituent exhibited greater activity. The ferric-reducing antioxidant power (FRAP) experiment revealed that complexes with hydroxyl substituents have high antioxidant potential. These complexes with IC50 values ranging from 0.86 to 2.47 mg mL-1 outperformed ascorbic acid, with an IC50 of 2.68 mg mL-1. The reported complexes bound to CT-DNA via intercalation mode with significant binding constants for complexes whose ligands bore the OH- substituent. Bovine serum albumin (BSA) binding affinity to the complexes ranges from moderate to high. The studied complexes with methyl substituents are promising anti-cervical cancer candidates.
Keywords: 3-pyridinyl Schiff bases, anion variation, anticancer, discrete Ag(I) complexes
INTRODUCTION
Imine represents one of the most extensively used organic compounds in various chemistry areas such as synthetic chemistry,1,2,3 catalysis,4 electrochemistry,5 materials science,6 and pharmaceutical chemistry.7,8,9 The main structural feature of an imine compound with general formula (R1-CH = N-R2) is the carbon-to-nitrogen double bond, where R represents a variable substituted alkyl or aryl group. The structure of Schiff bases is fundamental for their function in biological applications. Pyridinyl Schiff bases characterized by pyridinyl and a phenyl ring linked by an azomethine group have been of great interest because of their wide applications in various fields. It is known that free pyridinyl Schiff bases possess good biological activities, but when administrated as metal complexes, the bioactivity of the Schiff base is enhanced.10 Transition metals have gained a significant position in medicine because of the significant progress in using metallodrugs to treat several human diseases. Recent studies showed that transition metal complexes are potential biological agents such as antitumor,11 antiviral,12 anti-diabetes,13 antioxidant14 and antimicrobial.15 Among these transition metals, Ag(I) complexes have a promising future in medicine as therapeutic agents. This is related to Ag(I) complexes' exceptional chemical and biological features, such as variable chelating ability, high stability, high bio-activity at low concentrations, high selectivity and low toxicity to human beings.
Cancer is the world's second-largest cause of death. As a result, finding novel and more effective anticancer drugs with high cytotoxicity is critical. Over the years, various transition metal complexes as anticancer agents11,16,17,18 have been reported, but their usage has either been stopped or limited due to severe side effects, such as high toxicity to human beings, nephrotoxicity, neurotoxicity, ototoxicity, as haemolytic anaemia, drug resistance, poor selectivity and low bioavailability. Among the commonly used transition metals as anticancer agents, the literature has described Ag(I) complexes as one of the leading antitumor agents owing to their effective cytotoxicity against various tumour cells.
The evaluation of transition metal complexes' interactions with biomolecules, including DNA and proteins, is noted to have a key role in developing therapeutic agents. Since DNA contains all cellular information, the study of DNA interaction with a potential therapeutic agent that targets DNA is crucial. For example, The majority of anticancer medicines target DNA. In cancer cells, interactions between DNA and small molecules can cause damage, impede cell division, and lead to cell death. Ag(I) complexes have been recognized as good DNA binders. They can bind with DNA via different modes such as intercalation, groove and external electrostatic interaction. Likewise, the binding affinity of Ag(I) complexes with serum albumins have been intensively studied. A potential drug should possess a binding affinity to a carrier protein in the blood to be delivered to the expected target cells. Serum albumin, being the most important carrier protein in the blood, can bind with drug molecules and form a stable protein-drug complex affecting the absorption, distribution, activity and toxicity of such drugs. 19
Notably, in searching for new and effective antimicrobial agents to curtail the incessant resistance of microbes to antimicrobials, Ag(I) complexes have proven to be an alternative antimicrobial agent, as gathered from the literature.20,21,22,23 In like manner, many Ag(I) complexes with antioxidant properties24,25,26 have been developed. They can slow down or hinder the oxidation of biomolecules by terminating the free radicals' initiation or propagation reaction chains.
In continuation of our efforts in designing new Ag(I) complexes of pyridinyl Schiff bases as potential therapeutic agents, we report here the synthesis and characterization of pyridinyl functionalized Schiff bases obtained from the condensation reaction of 3-pyridinecarboxaldehyde with 2-aminophenol, 1-amino-2-fluorobenzene, and 4-methylaniline, respectively, and their Ag(I) complexes. The interaction of the pyridinyl Ag(I) complexes with DNA and Bovine serum albumin (BSA) and their antioxidant, antibacterial, and cytotoxicity assay were investigated.
EXPERIMENTAL
Materials and Methods
Local suppliers provided 3-pyridinecarboxaldehyde 99% (Aldrich, USA), 2-aminophenol >99% (Aldrich, USA), 4-methylaniline 99.5% (Aldrich, USA), 1-amino-2-fluorobenzene 99.5% (Aldrich, USA), calf-thymus DNA (CT-DNA), ethidium bromide 95% (Aldrich, USA), bovine serum albumin 99% (Aldrich, USA), >99% ethanol (Aldrich, USA), >99% diethyl ether (Aldrich, USA) and >99% DMSO-d6 (Merck, Germany). All of the chemicals were of analytical quality and were utilized as received, with the exception of the solvents, which were dried using traditional methods.
On a BRUKER 400 MHz spectrometer, 1H and 13C NMR spectra in DMSO-d6 and acetone-d6 were obtained. Chemical shift values in DMSO-d6 and acetone-d6 are presented in parts per million (ppm) relative to the solvent residual peaks; 2.5 and 2.05 ppm for 1H NMR and 39.5 and 29.4 ppm for 13C NMR, respectively. S stands for singlet, d for doublet, and m for multiplet in the splitting patterns of 1H NMR spectra, whereas J (in hertz) stands for the coupling constant. The infrared spectra were captured with an Agilent Cary 630 spectrometer, and the results are presented as percentage transmittances at various wavenumbers (cm-1), ranging from 4000 to 650 cm-1. Only molecular ions (M+) and significant fragmentation peaks were reported in the mass spectra, with intensities expressed as percentages of the base peak, using a Shimadzu LCMS-2020 instrument. Thermal-Scientific Flash 2000 CHNS/O analyzer was used to conduct elemental studies. The Stuart Scientific melting point device was used to determine all melting points. The Shimadzu UV-1800 UV-Vis Spectrophotometer was used to measure the absorption of CT-DNA and protein. A PerkinElmer LS 45 Fluorescence Spectrometer was used to record photoluminescence investigations of CT-DNA and protein.
Synthesis of pyridinyl Schiff bases L1-L3
Schiff bases (E)-2-((pyridin-3-ylmethylene)amino)phenol (L1), (E)-N-(2-fluorophenyl)-1-(pyridin-3-yl)methanimine (L2) and (E)-1-(pyridin-3-yl)-N-(p-tolyl)methanimine (L3) were synthesized using a slightly modified literature procedures.3,27,28 In the presence of glacial acetic acid, a hot solution of 3-pyridinecarboxaldehyde (1 mmol,~0.1 mL) in anhydrous ethanol (10 mL) was added to a hot solution of 2-aminophenol (1 mmol, 0.1 g), 1-amino-2-fluorobenzene (1 mmol, 0.1 mL), and 4-methylaniline (1 mmol, 0.11 g) respectively. The resultant reaction was refluxed at 80 °C for 4 h. Under reduced pressure, L1 and L3 solutions were evaporated to one-third of their original volume, and ether (10 mL) was added to each solution to produce a precipitate. Filtration was used to extract the precipitates, then rinsed with cold ether (10 mL x2) and recrystallized from ethanol. The L2 solution, on the other hand, was entirely evaporated under pressure, with the resulting oil being recrystallized from ethanol.
Synthesis of Ag(I) complexes of pyridinyl Schiff bases (1a-3a, 1b-3b and 1c-3c)
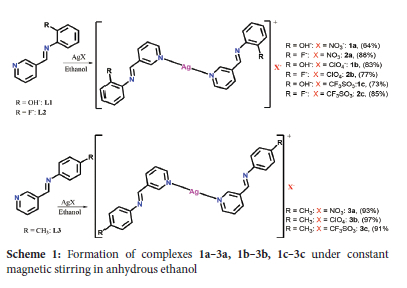
Nine Ag(I) complexes numbered 1a-3a, 1b-3b and 1c-3c were synthesized following our reported procedures.29 Under constant stirring in the dark, complexes 1a-3a, 1b-3b and 1c-3c were synthesized by dropwise addition of ethanol solution of each of L1-L3 (1 mmol) to an ethanol solution of AgNO3 (0.5 mmol, 0.10g), AgClO4 (0.5 mmol, ~0.10 g), and AgCF3SO3 (0.5 mmol, 0.13 g). For 6 hours, the reaction was carried out under N2 gas at room temperature. Vacuum filtering was later used to separate the precipitates that resulted. The precipitates were washed twice with cold ethanol, then twice with cold ether, before being dried in a vacuum. The isolated Ag(I) complexes were recrystallized by dissolving the complexes in dichloromethane and layering them with toluene.
[Ag(L1)2]NO3 1a
Colour: green, yield: 0.18 g, 64%, m.p 141-142 °C. 1H-NMR (400 MHz, DMSO-d6, 25 °C): δ ppm = 9.14 (1H, s, Hj-OH), 9.05 (1H, s, Hd-pyr), 8.80 (1H, s, He-C=N-), 8.69 (1H, s, Hc-pyr), 8.45 (1H, d, J = 7.82 Hz, Ha-pyr), 7.55 (1H, q, J = 4.74 Hz, Hb-pyr), 7.26 (1H, t, J = 7.88 Hz, Hf-ary), 7.11 (1H, m, Hi-ary), 6.92 (1H, d, J = 8.16 Hz, Hh-ary), 6.85 (1H, q, J = 7.57 Hz, Hg-ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 156.76 (C6- C=N-), 154.62 (C3-pyr), 151.45(C12-ary), 150.30 (C4-pyr), 137.35 (C7-ary), 136.16 (C1-pyr), 135.70 (C5-pyr), 128.05 (C10-ary), 119.59 (C2- pyr), 119.14 (C9-ary), 116.23 (C11-ary), 115.77 (C8-ary). FT-IR (cm-1): 3336, 3038, 1624, 1576. UV/Vis (acetonitrile): Amax 290, 347, 357 nm. MS: m/z Calcd. For [C24H20AgN4O2]: 504.32; found [Ag(L1)2+MeOH+K]+ 575 (11%), [Ag(L1-OH)]+ 288 (100%). Anal. Calcd. (%) for (C24H20AgN5O5): C, 50.90; H, 3.56; N, 12.37; found (%): C, 50.81; H, 3.42; N, 12.36.
[Ag(L2)2]NO3 2a
Colour: brown, yield: 0.25 g, 86%, m.p 171-172 °C. 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.09 (2H, d, Hd-pyr), 8.78 (2H, s, He-CH=N), 8.74 (2H, m, Hc- pyr), 8.38 (2H, Ha- pyr), 7.62 (2H, m, Hb-pyr), 7.31 (8H, m, Hf, g, h, i-ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 160.89 (C6- C=N-), 154.66 (C12- ary), 152.47 (C3- pyr), 150.66 (C4- pyr), 136.32 (C7- ary), 135.71 (C1- pyr), 131.50 (C5-pyr), 127.74 (c10- ary), 125.02 (C9- ary), 124.49 (C2- pyr), 124.36 (C8- ary). 116.26 (C11- ary). FT-IR (cm-1): (Ar-CH) 3254, (-C=N-) 1625, (pyridinyl) 1590, (C-F) 1488. UV/Vis (acetonitrile): λmax 235, 323 nm. MS: m/z Calcd. For [C24H18AgF2N4]: 508.30; found [Ag(L3) + H]+: 509 (100%), [AgL + Na+CH3CN + 2EtOH] +: 463 (67%), [AgL + CH3CN+H]+: 350 (20%). Anal. Calcd. (%) for (C24H18AgF2N5O3): C, 50.55; H, 3.18; N, 12.28; found (%): C, 50.39; H, 3.11; N, 12.10.
[Ag(L3)2]NO3 3a
Colour: white, yield: 0.26 g, 93%, m.p 159-160 °C. 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.07 (1H, s, Hd-pyr), 8.69 (2H, t, J = 1.72 Hz, 1.72 Hz, He-CH=N-, Hc-pyr), 8.32 (1H, m, Ha-pyr), 7.49 (1H, m, Hb-pyr), 7.23 (4H, s, Hf-ary), 2.36 (3H, s, Hg-CH3). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 157.33 (C6- C=N-), 151.91 (C3- pyr), 150.40 (C4- pyr), 148.29 (C7- ary), 135.94 (C10- ary), 135.23 (C1- pyr), 131.81 (C5- pyr), 129.71 (C9- ary , C12- ary), 124.19 (C2- pyr), 121.04 (C8- ary, C13- ary), 20.55 (C11- CH3). FT-IR (cm-1): (Ar-CH) 3054, (-C=N-) 1622, (pyridinyl) 1595, (C-CH3) 2895. UV/Vis (acetonitrile): λmax 287, 324 nm. MS: m/z Calcd. For [C26H24AgN4]: 500,37; found [Ag(L4)2 + H]: 501 (68%), [Ag(L4)] + MeOH + Na]: 359 (100%); [Ag(L4) + MeOH]: 336 (88%). Anal. Calcd. (%) for (C26H24AgN5O3): C, 55.53; H, 4.30; N, 12.45; found (%): C, 55.52; H, 4.27; N, 12.24.
[Ag(L1)2]ClO4 1b
Colour: brown, yield: 0.25 g, 83%, m.p 174-175 °C. 1H-NMR (400 MHz, DMSO-d6, 25 °C): δ ppm = 9.16 (2H, d, Hi-OH), 9.07 (2H, s, Hd-pyr), 8.83 (2H, s, H-C=N-), 8.70 (2H, q, J = 4.78 Hz, Hc-pyr), 8.47 (2H, d, J = 7.93 Hz, Ha-pyr), 7.57 (2H, q, J = 4.64 Hz, Hb-pyr), 7.28 (2H, q, J = 7.86 Hz, Hf- ary), 7.13 (2H, m, Hh-ary), 6.90 (4H, m, Hg- ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 157.24 (C6-C=N-), 155.07 (C3- pyr), 151.91 (C12- ary), 150.88 (C4- pyr), 137.81 (C7- ary), 136.57 (C1- pyr), 135.97 (C5- pyr), 128.45 (C10- ary), 120.01 (C2- pyr), 119.97 (C9- ary), 119.57 (C11- ary), 116.66 (C8- ary). FT-IR(cm-1); 3393, 1625, 1582, 1481, 1147, 1069. UV/Vis (acetonitrile): λmax 290, 348, 356 nm. MS m/z: Calcd. for [Ag(L1)2]: 504.32; found [Ag(L1)2+Na+2H] 529 (78%); [Ag(L1)2+2EtOH+CH3CN] 636 (34%); [Ag(L1)2+EtOH+H] 551 (17%). Anal. Calcd. (%) for [Ag(L1)2]ClO4. C, 47.74; H, 3.34; N, 9.28; found (%): C, 47.55; H, 3.28; N, 9.03.
[Ag(L2)2]ClO4 2b
Colour: white, yield: 0.23, 77%, m.p 250-251 °C, 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.09 (2H, s, Hd-pyr), 8.78 (2H, s, He-CH=N), 8.74 (2H, m, Hc- pyr), 8.37 (2H, m, Ha- pyr), 7.62 (2H, m, Hb- pyr), 7.31 (8H, m, Hf, g, h, i-ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 160.92 (C6- C=N-), 154.68 (C12- ary), 152.53 (C3- pyr), 150.71 (C4- pyr), 139.03 (C7- ary), 135.68 (C1- pyr), 131.50 (C5- pyr), 127.75 (C10- ary), 127.75 (C9- ary), 124.36 (C2- pyr, C8- ary). 116.07 (C11-ary). FT-IR (cm-1): (Ar-CH) 3070, (-C=N-) 1621, (pyridyl) 1591, (CF) 1490. UV/Vis (acetonitrile): λmax 235, 323 nm. MS: m/z Calcd. For [C24H18AgF2N4]: 508.30; found [Ag(L3)2 + H]+: 509 (97%), [Ag(L3)2 + CH3CN + H]+: 350 (21%). Anal. Calcd. (%) for (C24H18AgClF2N4O4): C, 47.43; H, 2.99; N, 9.22; found (%): C, 47.41; H, 2.86; N, 9.28.
[Ag(L3)2]ClO4 3b
Colour: white, yield: 0.29 g, 97%, m.p 205-206 °C. 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.06 (2H, d, J = 1.60 Hz, Hd-pyr), 8.70 (4H, m, He-CH=N-, Hc-pyr), 8.32 (2H, m, Ha-pyr), 7.57 (2H, m, Hb-pyr), 7.24 (8H, s, Hf-ary), 2.33 (6H, s, Hg-CH3). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 157.68 (C6- C=N-), 152.21 (C3- pyr), 150.70 (C4- pyr), 148.63(C7- ary), 136.26 (C10- ary), 135.48 (C1- pyr), 132.09 (C5- pyr), 130.03 (C9- ary , C12- ary), 124.47 (C2- pyr), 121.36 (C8- ary, C13- ary), 20.88 (C11- CH3). FT-IR (cm-1): (Ar-CH) 3074, (-C=N-) 1624, (pyridinyl) 1597, (C-CH3) 2908. UV/Vis (acetonitrile): λmax 289, 324 nm. MS: m/z Calcd. For [C26H24AgN4]: 500,37; found [Ag(L4)2 + H]: 501 (100%). Anal. Calcd. (%) for (C26H24AgClN4O4): C, 52.06; H, 4.03; N, 9.34; found (%): C, 52.00; H, 4.16; N, 9.27.
[Ag(L1)2]CF3SO3 1c
Colour: brown, yield: 0.24 g, 73%, Decomp.: 184-185 °C. 1H-NMR (400 MHz, DMSO-d6, 25 °C): δ ppm = 9.15 (2H, d, J = 1.67 Hz, Hj-OH), 9.08 (2H, d, J = 1.41 Hz, Hd-pyr), 8.81 (2H, s, He-CH=N-), 8.69 (2H, m, Hc-pyr), 8.47 (2H, m, Ha-pyr), 7.57 (2H, m, Hb-pyr), 7.26 (2H, m, Hf- ary), 7.12 (2H, m, Hh-ary), 6.88 (4H, m, Hg, i- ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 156.66 (C6- C=N-), 154.57 (C3- pyr), 151.41 (C12- ary), 150.27 (C4- pyr), 137.28 (C7- ary), 136.13 (C1- pyr), 135.71 (C5- pyr), 128.00 (C10- ary), 124.41 (C2- pyr), 124.05 (C9-ary), 119.47 (C11- ary), 116.18 (C8- ary). FT-IR(cm-1); 3396, 1627, 1582, 1483, 1147. UV/Vis (acetonitrile): λmax 290, 348, 359 nm. MS m/z: Calcd. for [Ag(L1)2]: 503.06 found [Ag(L-OH)2 +2CH3CN+Na] 575 (36%), [Ag(L1)2+2EtOH+Na] 619 (15%); [Ag(L1)2+CH3CN] 545 (16%). Anal. Calcd. (%) for (C25H20AgF3N4O5S): C, 45.96; H, 3.09; N, 8.58; found (%): C, 45.83; H, 3.05; N, 8.56.
[Ag(L2)2]CF3SO3 2c
Colour: white, yield: 0.28 g, 85%, Decomp.: 215-216 °C. 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.10 (2H, d, J = 1.80 Hz, Hd-pyr), 8.80 (2H, s, He-CH=N), 8.76 (2H, m, Hc- pyr), 8.38 (2H, Ha- pyr), 7.63 (2H, m, Hb- pyr), 7.33 (8H, m, Hf, g, h, i-ary). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 160.93 (C6- C=N-), 153.67 (C12- ary), 152.48 (C3- pyr), 150.68 (C4- pyr), 138.94 (C7- ary), 135.55 (C1- pyr), 131.45 (C5- pyr), 127.71 (C10- ary), 124.98 (C9- ary), 124.29 (c2- pyr), 121.40 (C2- pyr, C8- ary). 116.25 (C11- ary). FT-IR (cm-1): (Ar-CH) 3064, (-C=N-) 1623, (pyridyl) 1592, (C-F) 1495. UV/Vis (acetonitrile): λmax 235, 323 nm. MS: m/z Calcd. For [C24H18AgF2N4]: 508.30; found [Ag(L3)2 + H]+: 509 (25%), [Ag(L) + H]+: 290 (100%). Anal. Calcd.
(%) for (C25H18AgF5N4O3S): C, 45.68; H, 2.76; N, 8.52; found (%): C, 45.65; H, 2.76; N, 8.42.
[Ag(L3)2] CF3SO3 3c
Colour: white, yield: 0.30 g, 91%, m.p 246-247 °C. 1H-NMR (400 MHz, DMSO-d6, δ ppm): 9.07 (2H, d, J = 1.62 Hz, Hd-pyr), 8.71 (4H, t, He- CH=N-, Hc-pyr), 8.35 (2H, m, Ha-pyr), 7.61 (2H, m, Hb-pyr), 7.24 (8H, s, Hf-ary), 2.30 (6H, s, Hg-CH3). 13C-NMR (100 MHz, DMSO-d6, 25 °C): δ = 157.24 (C6- C=N-), 152.06 (C3- pyr), 150.50 (C4- pyr), 148.24 (C7- ary), 136.01 (C10- ary), 135.59 (c1- pyr), 131.94 (C5-pyr), 129.71 (C9- ary, C12- ary), 124.35 (C2- pyr), 121.06 (C8- ary, C13- ary), 20.55 (C11- CH3). FT-IR (cm-1): (Ar-CH) 3051, (-C=N-) 1625, (pyridyl) 1599, (C-CH3) 2927. UV/Vis (acetonitrile): λmax 291, 325 nm. MS: m/z Calcd. For [C26H24AgN4]: 500,37; found [Ag(L4)2 +L4+4]: 701 (42%), [Ag(L4)] + MeOH + Na]: 359 (100%); [Ag(L4) + 3MeOH]: 400 (26%). Anal. Calcd. (%) for (C27H24AgF3N4O3S): C, 49.94; H, 3.73; N, 8.63; found (%): C, 49.94; H, 3.64; N, 8.68.
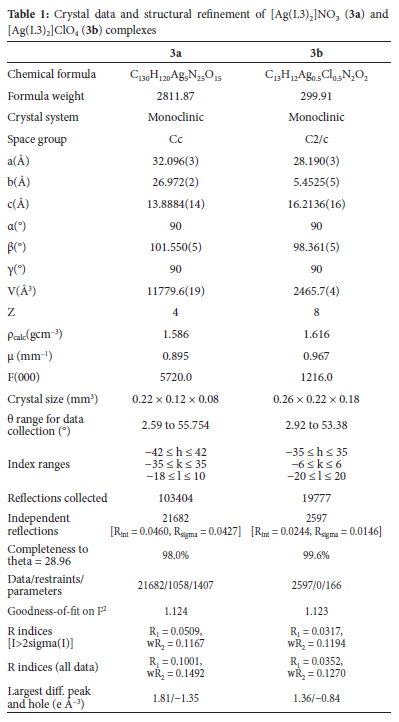
Single-crystal X-ray diffraction
On a Bruker Apex Duo diffractometer equipped with an Oxford Instruments Cryojet running at 100(2) K and an Incoatec microsource working at 30 W power, crystal evaluation and data gathering of [Ag(L3)2]NO3 (3a) and [Ag(L3)2]ClO4 (3b) were recorded. The results were collected using omega and phi scans with Mo K (= 0.71073) radiation at a crystal-to-detector distance of 50 mm. Outlier rejection, scan speed scaling, and standard Lorentz and polarization correction factors were used to reduce the data using the SAINT30 programme. The data were subjected to a SADABS31 semi-empirical multi-scan absorption adjustment.
The direct technique was used to solve and refine structures of 3a and 3b using the SHELXS32 program. ORTEP-333 system software was used to visualize the crystal structure information. Non-hydrogen atoms were refined isotropically initially, then anisotropically using SHELXL34 and a full-matrix least-squares approach based on F2. During refinement, a non-crystallographic inversion center was detected in the crystal lattice of 3a, and it was first suspected that the space group is centrosymmetric (C2/c), but the solution had much worse refinement statistics. Thus, an inversion twin was modelled and refined with a BASF value of 0.6(1). All hydrogen atoms were geometrically positioned, permitted to ride on their parent atoms, and isotropically purified. Table 1 lists the crystallographic data and structure refinement parameters for 3a and 3b.
In vitro antimicrobial studies
The sterile nutrient agar medium was made as described by Njogu et al. 201735 by dissolving 38 g of MHA in distilled water (1 L). The resulting nutrient agar medium was sterilized by autoclaving for 15 minutes at 121°C and cooled in a water bath to 45°C. The cooled agar media was put onto Petri dishes with a uniform 4 mm depth and then cooled to room temperature. Ag(I) complexes 1a-3a, 1b-3b, 1c-3c, as well as their ligands, were tested against six bacteria, including Salmonella typhimurium ATCC 14026, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 31488, Staphylococcus aureus ATCC 700699 (methicillin-resistant) and Staphylococcus aureus ATCC 25923. The nutrient broth was prepared by dissolving 1.3 g in 100 mL of distilled water and sterilized at 121 °C. Approximately 10 mL of the sterile nutritional broth was transferred to cotton wool-plugged test tubes and coated with aluminum foil. The nutritional broth in the test tubes was autoclaved for 15 minutes at 121 °C before being cooled to 37 °C. Following that, a single colony of the bacteria was isolated and put into a 10 mL sterile NB, which was then cultured at 37 °C for 18 hours in a shaking incubator. Each bacteria strain concentration was adjusted with sterile distilled water to a final concentration of 1.5 χ 108 cfu mL-1. The antibacterial activity of L1-L3 and 1a-3a, 1b-3b and 1c-3c was first screened by spotting 5 μL of each concentration of L1-L3 and 1a-3a, 1b-3b and 1c-3c on an MHA petri dish and incubating at 37 °C for 18 hours. By looking for a clear zone at the spotting site, the antibacterial activity was calculated. The minimum inhibitory concentration (MIC) of the active compounds with putative antibacterial activities was determined against six bacteria. Ten serial dilutions of L1-L3 and 1a-3a, 1b-3b, 1c-3c were performed, and the solution was further diluted 5 times where the compounds had lower MICs. The MIC of the compounds was measured by spotting 5 μL of each concentration of L1-L3 and 1a-3a, 1b-3b, 1c-3c on MHA plates and incubating them at 37 °C for 18 hours. These procedures were carried out three times to determine the exact lowest concentration of the complexes at which no observable bacterial growth could be detected following incubation. As a reference, Ciprofloxacin was employed, while 0.5% DMSO was used as a negative control.36
Antioxidant assay
The ferric reducing capacity of L1-L3 and 1a-3a, 1b-3b and 1c-3c was measured using Benzie and Strain method37 with slight modifications. Different concentrations ranging from 0.25 mg mL-1 to 2 mg mL-1 of L1-L3 and 1a-3a, 1b-3b and 1c-3c, as well as a standard (Ascorbic acid), were combined with 2.5 mL phosphate buffer (0.2 M) and 2.5 mL (1 percent w/v) potassium ferricyanide [K3Fe(CN)6]. The reaction mixture was acidified with 2.5 mL of trichloroacetic acid after 20 minutes of incubation at 50 °C (10%). After that, 1 mL of the acidified mixture was mixed with 1 mL of distilled water and 0.5 mL of iron (III) chloride (0.1%). The mixture's absorbance was then measured at 700 nm. Increased ferric reducing antioxidant power was shown by a decrease in absorbance. As shown in equation 1, the antioxidant power of the compounds can be expressed as a percentage of ferric reducing antioxidant power ascorbic acid equivalent.
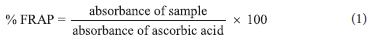
Cytotoxicity evaluation
The cytotoxicity of selected complexes; [Ag(L3)2]NO3 (3a), [Ag(L3)2] ClO4 (3b), [Ag(L3)2]CF3SO3 (3c) and Cisplatin as reference was evaluated on four human cell lines, human embryonic kidney 293 (HEK293), cervical cancer cell line (HELA), breast cancer cell line (MDA-MB231) and malignant human melanoma cell line (A375). Complexes 3a, 3b and 3c were selected for cytotoxicity based on our previous study38, where remarkable anticancer activity and high selectivity of Ag(I) complexes bearing the CH3 substituent were identified. The cytotoxicity experiment was performed using Abrahams et al. 20 1 839 methods, in which 1 χ 105 cells per mL of HEK293 (non-cancerous cell line), HELA, MDA-MB231 and A375 were seeded in 96-well microtiter plates and allowed to stabilize for 4 hours at 37 °C and 5% CO2. Then, through two-fold serial dilution, 3a, 3b, 3c and Cisplatin were added to the plate, resulting in eight final compound concentrations ranging from 100 to 0.781 μM in a total volume of 200 μL per well. The plate was then incubated for 72 h at 37 °C and 5% CO2. To each well, 20 μL CellTiter 96 AQueous One Solution (Promega, Madison, WI, USA) was added, and the plates were incubated for 4 h as previously described. After that, absorbance was measured using a multiplate reader at 490 nm (Molecular Devices, San Jose, CA, USA). EC50 values were obtained using GraphPad Prism 8 (2019) (GraphPad Software, San Diego, CA, USA) to estimate the concentration of each complex required to lower cell viability by 50%. The values are averages of at least three experiments (n = 3).
DNA binding assay
As described in the literature40,41 but with a slight modification, the calf thymus-DNA (CT-DNA) binding with L1-L3 and complexes 1a-3a, 1b-3b and 1c-3c was investigated using the absorption spectroscopy and fluorescence competitive displacement studies. The CT-DNA sodium salt was dissolved in phosphate buffer saline solution (PBS) pH 7.2 with continuous stirring overnight to make the DNA stock solution, filtered and kept at 4°C. UV-visible absorption with the absorption coefficient ε260 = 6600 M-1cm-1 was used to calculate the final concentration of the CT-DNA. The CT-DNA has a 1.93 ratio at 260 and 280 nm UV absorbance, showing it is clean and devoid of protein contamination. A constant concentration (50 μM) of the complexes in DMSO was also prepared. The DNA titration assay was carried out by titrating between 0 and 30 μM of CT-DNA in PBS against constant concentrations (50 μM) of L1-L3 and 1a-3a, 1b-3b and 1c-3c. The compounds-CT-DNA mixture was incubated for 10 mins before the absorbance was measured using Shimadzu UV-1800 UV-Vis Spectrophotometer. An equal amount of CT-DNA was added to the reference solution to eliminate CT-DNA absorbance.
The Wolfe-Shimer equation (Equation 2) was used to get the intrinsic binding constant, Kb, of the compounds:
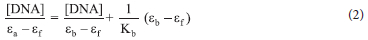
where [DNA], ea, eb, and ef represent DNA concentration, apparent, fully bound complex, and free complex extinction coefficients, respectively. The binding constant Kb of the compounds was calculated as the ratio of the slope to the intercept from the plot of [DNA]/(ea - ef) against [DNA].
Fluorescence competitive displacement study
In this study, a slightly mo dified metho d42 was used for the competitive fluorescence displacement assay. In a phosphate buffer of pH 7.2, a pre-treated 15 μΜ solution of ethidium bromide (EB) with 15 μΜ of CT-DNA was prepared and permitted to equilibrate for 30 mins. After that, different concentrations of the complexes (0-20 μΜ) were introduced at approximately 8 mins intervals for equilibration at room temperature, keeping CT-DNA-EB concentrations constant. The complexes' capacity to quench the fluorescence of CT-DNA bound EB was measured using a PerkinElmer LS 45 Fluorescence Spectrometer in the wavelength range of 530-700 nm at 25 °C with an excitation wavelength of 525 nm. The CT-DNA-EB fluorescence quenching of the complexes was evaluated using the Stern-Volmer Equation 3.
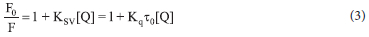
where F0 and F denote the relative fluorescence intensities of CT-DNA in the absence and presence of the quencher, Ksv denotes the SternVolmer quenching constant, [Q], quencher concentration, Kq denotes the bimolecular quenching constant, and r0 denotes the average lifetime of the fluorophore in the absence of the quencher, which in biomacromolecules is typically equal to 10-8 s.
Albumin binding assay using absorption spectroscopy and fluorescence quenching method
Bovine serum albumin (BSA) stock solution was made by dissolving an adequate amount of BSA in phosphate buffer saline solution (pH 7.2) at 25 °C. The BSA concentration was measured spectrophotometrically using ε280 = 44300 M-1cm-1 absorption coefficient. The complex stock solution was made by dissolving 1 mmol of the complexes in DMSO. The absorption titration experiment was performed by adding various concentrations of the complexes (0-10 μΜ) to a constant BSA solution (6 μΜ). After adding the concentration, the sample solution was incubated for 10 mins at 25 °C before recording the absorbance at λmax 280 nm. The intercept-to-slope ratio of 1/[A - A°] vs 1/[complex] linear curve was used to compute the binding constant Kb where A and A° denote BSA absorbance in the presence and absence of complexes, respectively.43,44
For the fluorescence quenching assay, a 1 μΜ BSA solution was titrated against various concentrations of the complexes (0-35 μΜ) and incubated at 25 °C for 8 mins. The wavelength of BSA was excited at 280 nm and emitted at 346 nm.45 The Stern-Volmer equation (Equation 4) was used to get the Ksv constant:
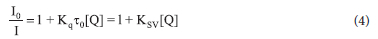
where I0 and I are the fluorescence intensities in the presence and absence of the quencher, respectively, and Kq Ksv, τ0, [Q] are the BSA quenching rate constant, dynamic quenching constant, the average lifetime of BSA without the quencher (τ0 = 10-8 s), and quencher concentration respectively.
RESULTS AND DISCUSSION
Synthesis and Characterization of L1-L3 and complexes 1a-3a, 1b-3b, 1c-3c
Ligands L1-L3 were synthesized by reacting 3-pyridinecarboxaldehyde with various substituted aromatic amines in anhydrous ethanol. Ligands L1, L2 and L3 were of fair to good yields at 72, 55, and 90%, respectively, and are all air-stable and soluble in ethanol, dichloromethane, dimethyl sulfoxide, acetonitrile, chloroform, and acetone. The Schiff bases were characterized using 1H NMR, 13C NMR, FT-IR, Mass and UV-Vis spectroscopy. In the IR spectra of ligands L1-L3, sharp peaks between 1619 and 1644 cm-1 were assigned to imine v(C=N), a feature of a Schiff base ligand. The 1H NMR spectra of L1, L2 and L3 in acetone-d6 exhibit singlets between 8.81, 8.72 and 8.69 ppm, respectively (Table S1) assigned to the imine proton (CH=N).
Complexes 1a-3a, 1b-3b and 1c-3c were prepared by reacting each ligand L1-L3 with AgNO3, AgClO4 and AgCF3SO3, respectively in 2:1 stoichiometry (L:M) in anhydrous ethanol. The complexes were produced in good to excellent yields ranging from 64 to 97% and were stable in the air. Various spectroscopic and analytical techniques were used to establish the complexes' formation and purity. The synthetic process for complexes 1a-3a, 1b-3b and 1c-3c is shown in Scheme 1.
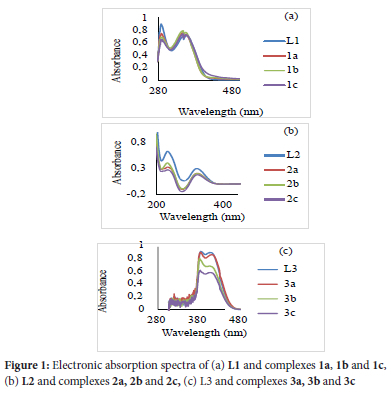
To follow ligand L1-L3 coordination mode, the 1H-NMR spectra of their protons near the donor atoms were compared to their respective complexes. In the 1H-NMR spectra of complexes 1a-3a, 1b-3b and 1c-3c, the integration values revealed the imine protons resonate between 8.72 and 8.81 ppm showing no notable chemical shift when compared to their free ligands L1, L2 and L3 chemical shift at 8.81, 8.72, 8.69 ppm respectively. On the other hand, the alpha protons on the pyridine ring of L1, L2 and L3 show chemical shifts at 8.64, 8.71 and 8.68 ppm, respectively, and on coordination (complexes 1a-3a, 1b-3b and 1c-3c) shifted downfield and resonated between 8.69 and 8.75 ppm suggesting the deshielding of the alpha protons in the pyridine ring by 0.01-0.07 ppm upon coordination to Ag(I). This suggests that L1-L3 coordinate to the Ag(I) ion via the Npy atom alone, forming discrete mononuclear complexes 1a-3a, 1b-3b and 1c-3c. This agrees with the reported Ag(I) complexes mode coordination of similar ligands.46 The formation of 1a-3a, 1b-3b and 1c-3c was also confirmed by correlating their FT-IR spectra with L1-L3 (Table S1). In the spectra of 1a-3a, 1b-3b and 1cv3c, the position of the -C=N-stretching frequencies remained nearly at the same frequency relative to their ligands, implying the non-involvement of the imine nitrogen atom in coordination with the Ag(I) ion. In contrast, the absorption bands attributed to the pyridinyl N atom between 1568 and 1581 cm-1 red-shifted between 1576 and 1595 cm-1 compared to their free ligand in 1a-3a, 1b-3b and 1c-3c. This implies that the coordination of L1-L3 to Ag(I) occurred through the Npy donor atom.47 The UV-Vis spectra of L1-L3, 1a-3a, 1b-3b and 1c-3c (Figure 1) were recorded in acetonitrile at room temperature. The spectra of L2 and L3 showed π-->π* transitions relative to the pyridinyl ring between 235 and 292 nm and imine n->π* transitions between 324 and 325 nm. L1 has three absorption bands at 288, 348 and 360 attributed to the pyridinyl ring π->π*, imine π->π* and imine n->π* nm transitions.48,49,50 The absorption patterns of complexes 2a, 2b, 2c, 3a, 3b and 3c are similar, having two absorption bands between 324 and 329 nm attributed to the imine n-π* transition and between 234 and 289 nm allied to the pyridinyl π-π* transition. Complexes 1a, 1b and 1c have three absorption bands. The first absorption band lies between 290 and 292 nm, the second between 348 and 349 nm, and the third is between 356 and 359 nm. These bands are due to the intra-ligand n-π* and π-π*and metal to ligand charge transfer transitions. Relative to their free ligands, the n->π* transitions redshifted in the absorption spectra of 1a, 1b, 1c, 2a, 2b and 2c, while in complexes 3a, 3b, and 3c, a slight blue shift was observed. In the mass spectra of 1a-3a, 1b-3b and 1c-3c, molecular ion peaks m/z = 504 for 1a, 1b and 1c correlating to [Ag(L1)2]+, while for 2a, 2b and 2c and 3a, 3b and 3c base peaks relating to [Ag(L2)2] + and [Ag(L3)2]+ were observed at m/z 509 and m/z 501 respectively (Table S2). The formation and purity of the reported structures in scheme 1 were confirmed using all the spectroscopic studies, as well as microanalysis (see ESI Table S1 and Table S2).
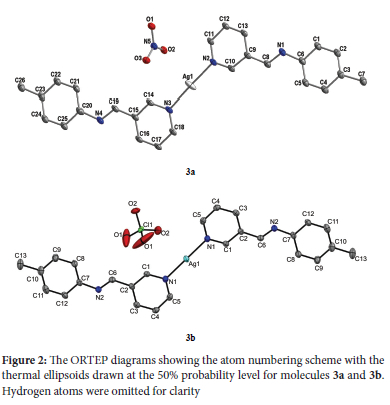
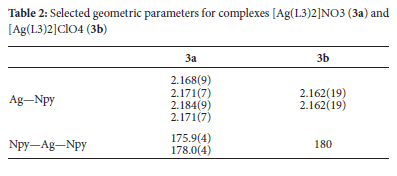
Crystal structures of complexes [Ag(L3)2]NO3 (3a) and [Ag(L3)2]ClO4 (3b)
The ORTEP diagrams and the atom numbering scheme for complexes 3a and 3b are shown in Figure 2. Complex 3a asymmetric unit has ten molecules of (E)-1-(pyridin-3-yl)-N-(p-tolyl)methanimine ligands, five Ag(I) ions and five nitrate anions, whereas complex 3b contains half a molecule of the cation [Ag(L3)]+ and half a molecule of the perchlorate anion. In both complexes, the cations are essentially planar in conformation. Around the Ag(I) ion, the complexes have a distorted linear geometry in which only the pyridine N atoms are involved in coordination with the Ag(I) center, with N-Ag-N bond angles ranging from 175.0(4) to 180° (Table 2). The anion molecules in both complexes are outside the coordination spheres. The angle around the Ag(I) centers in complexes 3a and 3b is similar to that of reported silver(I) complexes.51
In-vitro antimicrobial studies
Several antimicrobial agents have been used to treat infectious diseases52, yet multidrug-resistant bacteria pose a serious global health threat. The S. typhi, P. aeruginosa, E. coli, K. pneumoniae, Methicillin-resistant S. aureus and S. aureus bacteria selected in this study cause various human infections and diseases such as typhoid fever, gastroenteritis, urinary tract infections, respiratory system infections, soft tissue infections, neonatal meningitis, pneumonia, skin infections and endocarditis.53,54 These bacteria prove difficult to treat and tend to develop resistance to antimicrobial agents quickly, making antimicrobial agents less effective. Therefore, developing efficient novel molecules as a potential antimicrobial agent is crucial. The antibacterial activity of ligands; L1-L3, silver(I) complexes; 1a-3a, 1b-3b, 1c-3c, Ciprofloxacin (used as a control), and three silver salts was assessed against six bacteria strains using the minimum inhibitory concentration technique (MIC in μg mL-1). Table 3 lists the observed MIC values of the compounds.
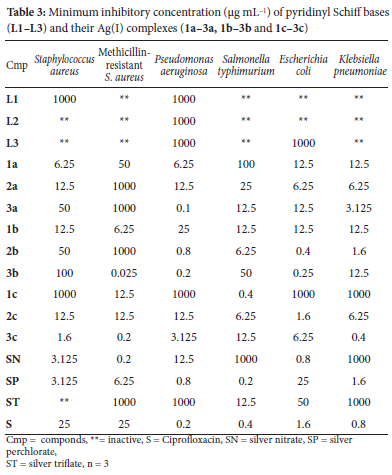
The antibacterial activities of the free ligands revealed L2 to be the least active ligand as it showed activity only against P. aeruginosa with 1000 μg mL-1 MIC value. L1, on the other hand, is selectively active against S. aureus and P. aeruginosa only, and L3 is active against P. aeruginosa and E. coli only. From the MIC values obtained for complexes 1a-3a, 1b-3b and 1c-3c against S. aureus, all the complexes except 1c, 2b, 3a and 3b showed better activity (1.6-12.5 μg mL-1) with respect to the reference standard (25 μg mL-1). 3c (with CH3 substituent) had the best activity than the reference standard and the metal salts against S. aureus. Similarly, almost all the complexes displayed significant activity against MRSA. MIC values between 0.025 and 12.5 μg mL-1 were recorded for all the complexes except 1a, 2a, 2b and 3a with 50-1000 μg mL-1 MIC against MRSA. These MIC results of the complexes against S. aureus and MRSA showed that most of the complexes had better activity than the reference standard towards gram-positive bacteria.
Towards the gram-negative bacteria, the complexes showed fair activities against P. aeruginosa with only 3a (with methyl substituent and NO3- as an anion) having a lower MIC (0.1 μg mL-1) than the reference standard (0.2 μg mL-1). 3b, also with methyl substituent, had the same MIC (0.2 μg mL-1) as the reference standard, and 2b (0.8 μg mL-1) had similar activities as the AgClO4 against P aeruginosa. Against S. typhi, only complex 1c (with OH substituent and CF3SO3-) had similar activity as the reference standard, while 2b and 2c, each with 6.25 μg mL-1 MIC had better activity than the AgNO3 and AgCF3SO3 salts against S. typhi. For the antimicrobial activities of the complexes against E. coli, 2b and 3b displayed better activity, with each of their MIC values (0.25-0.4 μg mL-1) lower than the reference standard (1.6 μg mL-1). 2c had the same MIC as the standard against E. coli. The activity of 3c with methyl substituent (0.4 μg mL-1) is double that of the standard (0.8 μg mL-1) against K. pneumoniae, while the activity of 2b (with F-) against K. pneumoniae is the same as that of AgClO4. Each of the complexes with better antimicrobial activity has either CH3 or F- substituent. This shows that F- and CH3 substituents and ClO4- and CF3SO3- anions play a role in the complexes' antimicrobial activity.
In general, all the complexes were fairly active against the tested bacteria strains in comparison with the standard, with 3c (with methyl substituent) somewhat showing notable antimicrobial activity against all the bacteria tested. Significant antimicrobial activity of compounds with methyl substituents 55,56 have been reported.
Overall, the coordination of ligands L1-L3 to the silver(I) ion impacted their antibacterial activity, as proven by the MIC values recorded. The improved antibacterial activity of the studied complexes can be linked to the mechanism of action of the silver(I) ion, where the silver(I) ion is slowly released from the ligand upon entry into the bacteria cell to interact with nucleic acids, protein thiol groups and several vital enzymes. This leads to protein denaturation, membrane malfunction, cell division blockage, and consequently, inhibiting the organism's growth.35 Chelation, known for the promotion of π-electron delocalization along a chelate ring and complexes lipophilicity increment, could have encouraged the complexes' absorption into the bacterial membranes and inhibited the metal-binding sites in microbial enzymes.57 The impact of the perchlorate and triflate anions, as well as the F- and CH3 substituents on the complexes' antibacterial activity, were also identified, which correlates with that of the literature.55,56 Relative to MIC values between 4 and 256 μg mL-1 of similar Ag(I) complexes reported,58,59,60 the Ag(I) complexes in this study demonstrated better antibacterial activity with MIC values between 0.2 and 100 μg mL-1.
Antioxidant study
Synthetic antioxidants can slow down or stop the damage to DNA, RNA and protein substrates caused by free radicals; they can also be used for stabilizing organic materials against degradation, such as additives in polymer formulations.61 The antioxidant activity of L1-L3 and 1a-3a, 1b-3b and 1c-3c were studied by the ferric reducing antioxidant power (FRAP) method. The FRAP assay is based on an antioxidant's capacity to reduce Fe(III) tripyridyltriazine complex to Fe(II) tripyridyltriazine complex.62 The compounds' 50% inhibitory concentrations (IC50 in mg mL-1) are used to express the antioxidant activity of the compounds. Ascorbic acid was utilized as a control, and its IC50 value was 2.68 mg mL-1. The IC50 values for L1-L3 and complexes 1a-3a, 1b-3b and 1c-3c are displayed in Table 4, and Figure 3 displays a histogram of the free radical scavenging activity vs. conc. (mg mL-1).
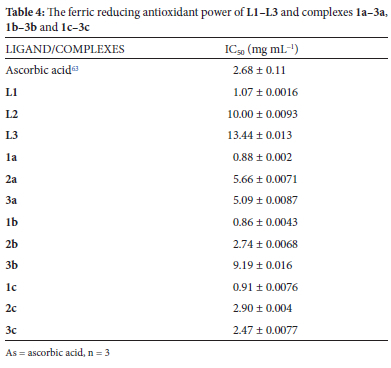
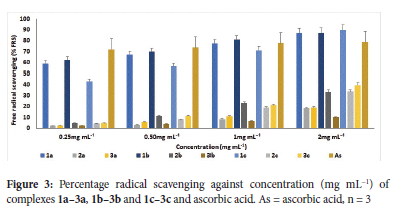
Ligands L1, L2 and L3 had 1.07, 10.00 and 13.44 mg mL-1 IC50, respectively, depicting L1 (with a hydroxyl group) as a good antioxidant compared to the reference IC50 of 2.68 mg mL-1. A further evaluation of the complexes' antioxidant activity revealed the complexes displayed higher antioxidant activities than the free ligands. Among all the complexes, 1a, 1b, 1c, and 3c exhibited significant antioxidant activity, with IC50 values ranging from 0.86 to 2.47 mg mL-1 (Table 4). These complexes demonstrated greater activity than ascorbic acid, whose capacity to reduce Fe(III) to Fe(II) is shown by an IC50 value of 2.68 mg mL-1. Most of these complexes with notable antioxidant activity have a phenolic ligand, which could have influenced their antioxidant property. This observation can be compared with El Jemli et al. 201664 reports, where phenolic compounds were identified with high antioxidant activity.
Anticancer studies
Transition metal complexes have gained more recognition as promising anticancer agents than platinum-based complexes previously employed. Therefore, this study used an MTT cell viability assay to ascertain the anticancer effects of complexes 3a, 3b, 3c and Cisplatin against HEK293, A375, HELA and MDA-MB231. The concentration of the complexes necessary to kill 50% of the cells and the selectivity index (SI) was determined and tabulated in Table 5. The results indicate that 3a, 3b, and 3c showed antiproliferative effects on HELA with EC50 values of 44.21 ± 1.87, 32.14 ± 3.91 and 28.07 ± 3.09 μM, respectively, better than Cisplatin (EC50 > 50 μM). The impact of the methyl group of the complexes on their remarkable cytotoxicity against HELA can be related to that of Sreelatha et al. 201465 who reported the significance of the methyl group in the cytotoxicity of naphthoquinone amide derivatives towards HELA. The type of counter ion present in the complex was also observed to have influenced their cytotoxicity as 3c (with CF3SO3- anion) had better cytotoxicity, followed by 3b (with ClO4- anion) and 3a (with NO3-anion) against the HELA. While 3a and 3b showed poor cytotoxicity when compared to Cisplatin (EC50 = 22.68 ± 4.89 μM) against MDA-MB231, the concentration of 3c (EC50 = 23.30 ± 3.82 μM) used to kill 50% of the cells was almost similar to that of Cisplatin. Against A375, 3a, 3b and 3c with EC50 between 33.73 ± 5.83 and 62.02 ± 7.23 μM showed low cytotoxicity when compared to Cisplatin with 3.4 ± EC50 value. The selectivity index of the complexes was determined as the quotient obtained by dividing the HEK293 EC50 value by the EC50 value of the respective cancer cell lines. In comparison to Cisplatin, all the complexes displayed poor selectivity for A375, HELA and MDA-MB231 cancer cells, according to the SI data in Table 5. This indicates that the complexes cannot selectively inhibit the growth of the cancer cells without attacking the normal cell.
DNA binding studies
In designing potential bioactive drugs, the interaction of such drug molecules with DNA is important since DNA is the primary target for most biological agents. The electronic absorption titration was used to follow the interactions of L1-L3, 1a-3a, 1b-3b and 1c-3c with CT-DNA; significant absorption bands between 274 and 277 nm attributed to the π -> π* transition energy were observed for the compounds. The absorption spectra of the compounds further showed a hypochromic shift between 6.34 and 58.62% and a bathochromic shift between 2 and 5 nm except for 1a, 3c, 4b and 4c with hypochromic and slight hypsochromic shifts. The hypochromic shift indicates the compounds bind in a non-covalent intercalative fashion to the CT-DNA double helix.66 The hypochromic absorption spectra of the compounds in the presence and absence of CT-DNA are displayed in Figures 4a and S49-S59 (see ESI).
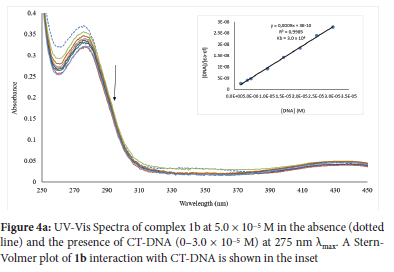
The "Kb" representing the binding constants of the compounds was determined using the Wolfe-Shimer equation (Eq. ii) by finding the ratio of the slope to the intercept from the plot of [DNA]/(εa - εf) versus [DNA] to compare quantitatively the affinity of L1-L3, 1a-3a, 1b-3b and 1c-3c toward CT-DNA. The binding constant of ligands L1-L3 was found to be between 1.33 χ 104 and 2.85 χ 105, while 1a-3a, 1b-3b and 1c-3c were found to be between 2.33 χ 105 and 3.00 χ 106 M-1(Table 6). The Kb value is proportional to the compounds' binding affinity and intercalative strength when it binds to DNA. Therefore, the ligands have poor interactions with CT-DNA not until their coordination to Ag(I) ion, which improves their binding affinity. Comparing the Kb value of the complexes, 1b (3.00 χ 106 M-1) had the highest binding affinity (Figure 4a), followed by 1c with 1.75 χ 106 M-1.
The strong binding affinity of these two complexes (1b and 1c) to CT-DNA could have been related to the hydroxyl group present in the complexes, similar to that of the literature.67
Fluorescence competitive displacement studies
The binding mode of 1a-3a, 1b-3b and 1c-3c to CT-DNA was further ascertained using the fluorescence competitive displacement experiments. It is worth mentioning that L1-L3 were exempted from this analysis since their CT-DNA binding affinity is lower than that of the complexes. The fluorescence competitive displacement investigation is based on the test complexes displacing ethidium bromide (an intercalative agent) from the CT-DNA-EB adduct. While performing the experiment, the gradual introduction of 1a-3a, 1b-3b and 1c-3c to ethidium bromide incubated with CT-DNA resulted in a significant reduction in the CT-DNA-EB fluorescence intensity at 605 nm (Figures 4b and S60-S67). This means the complexes could displace CT-DNA-bound EB and bind to CT-DNA at the intercalation site, thus validating the complexes binding to CT-DNA via intercalation mode.
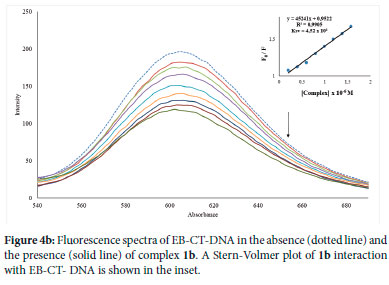
The quenching strength, Ksv, was then computed from the slope of the linear quenching plot of Fo/F versus the concentration of the complexes (Equation 3) to measure the relative binding interaction of the complexes with CT-DNA. The Ksv values for 1a-3a, 1b-3b and 1c-3c ranged from 1.61 to 4.52 x 104 M-1 (Table 6), demonstrating their high affinity for CT-DNA. The maximum binding affinity to CT-DNA was found in complex 1b (Figure 4b), which corresponds to the complex with the highest intrinsic Kb constant (Figure 4a). The quenching of the fluorescence intensity of EB-CT-DNA adduct by the complexes was further utilized to evaluate the quenching mechanism (Kq), binding constant (Kbin) and the binding sites (n). The quenching mechanism, which could either be static or dynamic, due to the obtained Stern-Volmer linear plots of the complexes (inset Figures 4b and S60-S67), can be evaluated from Equation 5.

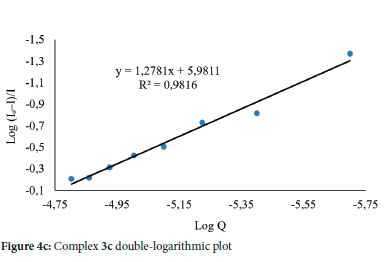
Ksv and Kq represent the Stern-Volmer quenching constant and bimolecular quenching constant, respectively, while τ0 represents an average fluorophore lifetime in the absence of the quencher, usually 10-8 s in biomacromolecules. The Kq of the complexes was determined to be between 1.61 and 4.52 x 1012 M-1S-1 (Table 6), which is higher than the maximal diffusion collision quenching rate constant of many biological macromolecule quenchers (2 x 1010 M-1S-1).68 The range of the Kq value obtained indicates that the quenching by a-3a, 1b-3b and 1c-3c was caused by a static quenching mechanism and that complexes formed between quenching and fluorescent molecules are stabilized at the ground state.69 The Kbin of complexes a-3a, 1b-3b and 1c-3c with CT-DNA were calculated using the double-logarithmic (Equation 6), where a plot of log F0 - F/F vs log [Q] of complexes 1a-3a, 1b-3b and 1c-3c yielded a straight line with a slope of n and y-axis intercept of log Kbin (Figures S68-S75)

The relative fluorescence intensities of CT-DNA in the absence and presence of the quencher, respectively, are represented by F0 and F. At the same time, [Q] is the quencher concentration, Kbin is the binding constant, and n is the number of binding sites. The values of n for complexes a-3a, 1b-3b and 1c-3c are all close to 1, indicating that all complexes bind to one reactive site on the CT-DNA, as shown in Table 6. With a binding constant (Kbin) equal to 9.57 x 105 M-1, Complex 3c (Figure 4c) had the highest binding affinity to CT-DNA.
Protein binding studies
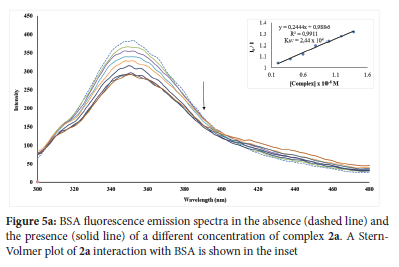
Drug molecules are transported well by proteins found in blood plasma. Therefore, the mechanism of protein interactions with a potential drug plays a fundamental role in understanding such drug pharmacokinetics and pharmacodynamics.70 The protein standard in this work was BSA, which was utilized to understand the pharmacokinetics and pharmacodynamics of a-3a, 1b-3b and 1c-3c. The interaction between a-3a, 1b-3b and 1c-3c and BSA were explored by fluorescence and electronic absorption spectroscopy. For the fluorescence assay, the excitation of BSA at 280 nm resulted in its emission between 300 and 400 nm, which can be attributable to the amino acid residues such as tyrosine (Tyr), tryptophan (Trp) and phenylalanine (Phe).71 As observed in the emission spectra of the compounds, the incremental addition of 2a, 2b and 2c to a constant concentration of BSA led to hypochromic shifts between 15.99 and 23.59% (an indication of complex formation) with the fluorescence intensity of the BSA around 350-360 nm with a slight redshift between 1 and 3 nm in the maximum emission. These findings suggest a complex formation between the BSA and studied compounds leading to the quenching of the BSA. The emission spectra of BSA in the presence of 2a, 2b and 2c are shown in Figure 5a and Figures S79 and S82. 2a (Figure 5a) quenched the intrinsic fluorescence of BSA more. BSA's fluorescence intensity was not quenched by complexes 1a, 1b, 1c, 3a, 3b and 3c. The presence of either OH- or CH3 in 1a, 1b, 1c, 3a, 3b and 3c could have limited their ability to access the protein's binding site buried in the hydrophobic environment. Non-binding interaction of BSA with similar substituents has been reported by our group.72 The possible quenching mechanism of the BSA fluorescence by 2a, 2b and 2c was determined by the Stern-Volmer Equation (Equation 4). The slope of the linear regression on the Stern-Volmer plot of I0/I vs [Q] yielded the Ksv values for the complexes, and the Kq value was determined using the equation:
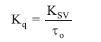
The Ksv and Kq values were found to be between 1.62 x 104 and 2.44 χ 104 Μ-1 and 1.62 χ 1012 and 2.44 χ 1012 Μ-1, respectively. The calculated Kq values were higher than the biomolecules' maximum scatter collision-quenching constant (2 χ 1010 Μ-1 S-1), indicating that the complexes' BSA quenching mechanism is static. Scatchard Equation vii can be used to calculate the binding constant (Kbin) and the number of binding sites (n) for static quenching:

The slope and intercept of the double logarithm regression of log(I0 - versus log[Q] (Figure 5b and Figures S84-S85) can be used to compute n and Kbin respectively. The Kbin values indicate that BSA has a modest affinity for the complexes. The maximum binding affinity for BSA was found in Complex 2a (Figure 5b). The number of the binding site (n) for complexes 2a, 2b and 2c are about one, implying that one molecule of each complex binds to BSA. The results obtained in this study reveal moderate interactions between these Ag(I) complexes and BSA, which occur through hydrophobic force and static mechanism.
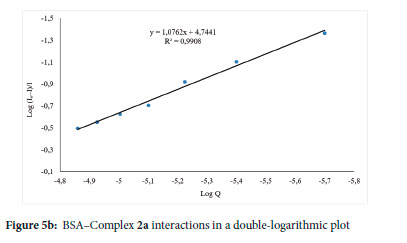
In the electronic absorption spectroscopy titration, each addition of complexes 1a-3a, 1b-3b and 1c-3c to a constant BSA concentration led to an increase in the BSA λmax at ~280 nm with slight redshifts (Figure 5c and Figures S86-S93). The intercept-to-slope ratio of the linear curve of a plot of 1/[A - A0] vs 1/[complex] yielded the Kb constant in this case. The complexes' Kb value falls between 2.30 χ 104 and 3.67 χ 105 Μ-1 (Table 5), which is similar to Anjomshoa et al. 20 1 443 binding constant values between 104 and 106 M-1. The maximum binding affinity for BSA was found in complex 3b (Figure 5c). A similar literature report73 can be linked to the effect of 3b ligand substituents on their binding affinity to BSA.
CONCLUSION
Nine sets of mononuclear discrete Ag(I) complexes of aryl substituted (E)-N-phenyl-1-(pyridin-3-yl)methanimine were synthesized, characterized and bio-evaluated for antibacterial, antioxidant and anticancer activities as well as their binding with biomolecules. Based on the spectral evidence, the ligands coordinated to the Ag(I) center in a 2:1 stoichiometry (L:M) through the nitrogen atom of the pyridine only, with complexes 3a and 3b having a linear geometry around the Ag(I) center as obtained from the single crystal X-ray diffraction analysis. The biological evaluations of the complexes reveal their structural activity relationship. From the antibacterial screening, complexes possessing either CH3 or F- substituent showed remarkable activity against the tested microorganisms. The remarkable impact of the ClO4- and CF3SO3- anions on the complexes' antibacterial activities was also observed. The results of the antioxidant study by the FRAP method showed that complexes with OH- substituent exhibit good antioxidant activity with reference to ascorbic acid. UV-Vis absorption and fluorescence spectroscopy were used to investigate the complexes' interactions with CT-DNA and BSA. The binding studies of the Ag(I) complexes with CT-DNA suggested their intercalative mode of interaction. The high Kb constant value revealed that all the complexes had a high binding affinity for CT-DNA. All the complexes exhibit significant binding affinity to BSA, and the fluorescence quenching of the BSA by the Ag(I) complexes were initiated by a static mechanism. The moderate to high binding affinities of the Ag(I) complexes with CT-DNA and BSA denotes that the novel Ag(I) complexes can bind to DNA and serum albumin. The complexes showed potential anti-cervical cancer activity. Hence, the Ag(I) complexes are potential therapeutic alternatives since their remarkable antimicrobial, antioxidant and anticancer activities were recorded.
CONFLICT OF INTEREST
No potential conflict of interest was reported by the authors.
AUTHORS CONTRIBUTION
All authors contributed to the preparation of this manuscript. BO provided the resources and supervised the project. AAA synthesized, characterized the metal complexes and carried out the biological studies of the complexes. KO and VFS did the antioxidant analysis. AAA wrote the original draft. BO and Md.SI reviewed the manuscript.
FUNDING
The authors are grateful for financial support from the National Research Foundation of South Africa (Grant number: 119342).
ACKNOWLEDGMENTS
The authors appreciate the help and use of the facilities from the University of KwaZulu-Natal, South Africa.
SUPPLEMENTARY MATERIAL
Supplementary information for this article is provided in the online supplement.
ORCID IDs
Adesola A. Adeleke: https://orcid.org/0000-0001-9287-5960
Md. Shahidul Islam: https://orcid.org/0000-0003-0309-9491
Kolawole Olofinsan: https://orcid.org/0000-0002-2987-0996
Veronica F. Salau: https://orcid.org/0000-0002-3796-4992
Bernard Omondi: https://orcid.org/0000-0002-3003-6712
REFERENCES
1. Vhanale BT, Deshmukh NJ, Shinde AT. Synthesis, characterization, spectroscopic studies and biological evaluation of schiff bases derived from 1-hydroxy-2-acetonapthanone. Heliyon. 2019;5(11):e02774. https://doi.org/10.1016/j.heliyon.2019.e02774 [ Links ]
2. Adeleke A, Omondi B. Crystal structure of a chiral sec-amine, 4-chloro-n-(1-(pyridin-2-yl)ethyl)aniline. Molbank. 2022;2022(1). https://doi.org/10.3390/m1335 [ Links ]
3. Adeleke A, Zamisa SJ, Omondi B. Crystal structure of 4-(1-phenylimidazo[1,5-a]pyridin-3-yl)benzoic acid (C20H14N2O2). Zeitschrift für Kristallographie - New Crystal Structures. 2019;234(6):1157-1159. https://doi.org/10.1515/ncrs-2019-0300 [ Links ]
4. Gupta K, Sutar AK. Catalytic activities of schiff base transition metal complexes. Coord Chem Rev. 2008;252(12-14):1420-1450. https://doi.org/10.1016/j.ccr.2007.09.005 [ Links ]
5. Oiye E, Ribeiro MFM, Katayama JMT, Tadini MC, Balbino MA, Eleoterio IC, Magalhaes J, Castro AS, Silva RSM, da Cruz Junior JW, Dockal ER, de Oliveira MF. Electrochemical sensors containing schiff bases and their transition metal complexes to detect analytes of forensic, pharmaceutical and environmental interest. A review. Crit Rev Anal Chem. 2019;49(6):488-509. https://doi.org/10.1080/10408347.2018.1561242 [ Links ]
6. Zhang J, Xu L, Wong W-Y. Energy materials based on metal schiff base complexes. Coord Chem Rev. 2018;355:180-198. https://doi.org/10.1016/j.ccr.2017.08.007 [ Links ]
7. Chaturvedi D, Kamboj M. Role of schiff base in drug discovery research. Chem Sci J. 2016;7(2):e114. https://doi.org/10.4172/2150-3494.1000e114 [ Links ]
8. Kajal A, Bala S, Kamboj S, Sharma N, Saini V. Schiff bases: A versatile pharmacophore. J Catal. 2013;2013:1-14. https://doi.org/10.1155/2013/893512 [ Links ]
9. Zamisa S, Ngubane NP, Adeleke AA, Jonnalagadda SB, Omondi B. Classical intermolecular hydrogen bonding motifs of heterocyclic rac-2-amino-3-carbonitrile derivatives: Linking hirshfeld surface analysis, ct-DNA binding affinity, and molecular docking. Cryst Growth Des 2022;22(10):5814-5834. https://doi.org/10.1021/acs.cgd.1c01514 [ Links ]
10. Mendu P, Kumari CG, Ragi R. Synthesis, characterization, DNA binding, DNA cleavage and antimicrobial studies of schiff base ligand and its metal complexes. J Fluoresc. 2015;25(2):369-378. https://doi.org/10.1007/s10895-015-1520-6 [ Links ]
11. Nandanwar S, Kim HJ. Anticancer and antibacterial activity of transition metal complexes. ChemistrySelect. 2019;4(5):1706-1721. https://doi.org/10.1002/slct.201803073 [ Links ]
12. Pal M, Musib D, Roy M. Transition metal complexes as potential tools against sars-cov-2: An in silico approach. New J Chem. 2021;45(4):1924-1933. https://doi.org/10.1039/D0NJ04578K [ Links ]
13. Levina A, Lay PA. Metal-based anti-diabetic drugs: Advances and challenges. Dalton Trans. 2011;40(44):11675-11686. https://doi.org/10.1039/c1dt10380f [ Links ]
14. Yusuf T, Akintayo DC, Oladipo SD, Adeleke AA, Olofinsan K, Vatshaa B, Mabuba N. The effect of structural configuration on the DNA binding and in vitro antioxidant properties of new copper(II) N2O2 schiff base complexes. New J Chem. 2022:1-13. https://doi.org/10.1039/D2NJ01477G [ Links ]
15. Khan S, Nami SAA, Bhat SA, Kareem A, Nishat N. Synthesis, characterization and antimicrobial study of polymeric transition metal complexes of Mn(II), Co(II), Ni(II), Cu(II) and Zn(II). Microb Pathog. 2017;110:414-425. https://doi.org/10.1016/j.micpath.2017.07.008 [ Links ]
16. Ndagi U, Mhlongo N, Soliman ME. Metal complexes in cancer therapy - an update from drug design perspective. Drug Des Devel Ther. 2017;11:599-616. https://doi.org/10.2147/DDDT.S119488 [ Links ]
17. Hussain A, AlAjmi MF, Rehman MT, Amir S, Husain FM, Alsalme A, Siddiqui MA, AlKhedhairy AA, Khan RA. Copper(II) complexes as potential anticancer and nonsteroidal anti-inflammatory agents: In vitro and in vivo studies. Sci Rep. 2019;9(1):5237. https://doi.org/10.1038/s41598-019-41063-x [ Links ]
18. Ghdhayeb M, Haque RA, Budagumpi S, Khadeer Ahamed MB, Abdul Majid AMS. Mono- and bis-N-heterocyclic carbene silver(I) and palladium(II) complexes: Synthesis, characterization, crystal structure and in vitro anticancer studies. Polyhedron. 2017;121:222-230. https://doi.org/10.1016/j.poly.2016.09.065 [ Links ]
19. Tesseromatis C, Alevizou A. The role of the protein-binding on the mode of drug action as well the interactions with other drugs. Eur J Drug Metab Pharmacokinet. 2008;33(4):225-230. https://doi.org/10.1007/BF03190876 [ Links ]
20. Karata? M, Günal S, Mansur A, Alici B, Qetinkaya E. Synthesis and antimicrobial properties of cycloheptyl substituted benzimidazolium salts and their silver(I) carbene complexes. Heterocycl Comm. 2016;22(6):357-361. https://doi.org/10.1515/hc-2016-0105 [ Links ]
21. Roymahapatra G, Mandal SM, Porto WF, Samanta T, Giri S, Dinda J, Franco OL, Chattaraj PK. Pyrazine functionalized Ag(I) and Au(I)-NHC complexes are potential antibacterial agents. Curr Med Chem. 2012;19(24):4184-4193. https://doi.org/10.2174/092986712802430090 [ Links ]
22. Gok Y, Akkoc S, Erdogan H, Albayrak S. In vitro antimicrobial studies of new benzimidazolium salts and silver N-heterocyclic carbene complexes. J Enzyme Inhib Med Chem. 2016;31(6):1322-1327. https://doi.org/10.3109/14756366.2015.1132210 [ Links ]
23. Melaiye A, Youngs WJ. Silver and its application as an antimicrobial agent. Expert Opin Ther Pat. 2005;15(2):125-130. https://doi.org/10.1517/13543776.15.2.125 [ Links ]
24. Wu H, Yuan J, Bai Y, Pan G, Wang H, Kong J, Fan X, Liu H. Synthesis, structure, DNA-binding properties and antioxidant activity of silver(I) complexes containing v-shaped bis-benzimidazole ligands. Dalton Trans. 2012;41(29):8829-8838. https://doi.org/10.1039/c2dt30512g [ Links ]
25. Slimani I, Mansour L, Abutaha N, Harrath AH, Al-Tamimi J, Gürbüz N, Özdemir I, Hamdi N. Synthesis, structural characterization of silver(I)-NHC complexes and their antimicrobial, antioxidant and antitumor activities. J King Saud University - Science. 2020;32(2):1544-1554. https://doi.org/10.1016/j.jksus.2019.12.010 [ Links ]
26. Tang X, Qu Y, Zhao K, Wang C, Wu Y, Mao S, Wu H. Synthesis, crystal structure, fluorescence and antioxidant properties of a binuclear Ag(I) complex based on the spph3 ligand. J Chem Res. 2019;43(5-6):184-188. https://doi.org/10.1177/1747519819857503 [ Links ]
27. Jehdaramarn A, Pornsuwan S, Chumsaeng P, Phomphrai K, Sangtrirutnugul P. Effects of appended hydroxyl groups and ligand chain length on copper coordination and oxidation activity. New J Chem. 2018;42(1):654-661. https://doi.org/10.1039/c7nj03113k [ Links ]
28. Adeleke A, Zamisa SJ, Omondi B. Crystal structure of dichlorido-bis((E)-2-((pyridin-4-ylmethylene)amino)phenol)zinc(II), C24H20Cl2N4O2Zn. Z Kristallogr NCS. 2020;235(3):625-628. https://doi.org/10.1515/ncrs-2019-0863 [ Links ]
29. Adeleke A, Zamisa SJ, Omondi B. Ag(I) complexes of imine derivatives of unexpected 2-thiophenemethylamine homo-coupling and bis-(E)-N-(furan-2-ylmethyl)-1-(quinolin-2-yl)methanimine. Molbank. 2021;2021(2):1-9. https://doi.org/10.3390/m1235 [ Links ]
30. SAINT BVA. Data reduction software. Bruker AXS Inc, Madison, Wisconsin, USA. 2009. [ Links ]
31. Bruker A. Saint and sadabs. Bruker AXS Inc, Madison, Wisconsin, USA. 2009. [ Links ]
32. Sheldrick G. Crystal structure solution with SHELXT. Acta Crystallogr A. 2015;71:3-8. https://doi.org/10.1107/S2053273314026370 [ Links ]
33. Farrugia L. WinGX and ORTEP for Windows: An update. J Appl Cryst. 2012;45(4):849-854. https://doi.org/10.1107/S0021889812029111 [ Links ]
34. Sheldrick G. Crystal structure refinement with SHELXL. Acta Crystallogr C Struct Chem. 2015;71(Pt 1):3-8. https://doi.org/10.1107/S2053229614024218 [ Links ]
35. Njogu E, Omondi B, Nyamori VO. Silver(I)-pyridinyl Schiff base complexes: Synthesis, characterisation and antimicrobial studies. J Mol Struct. 2017;1135:118-128. https://doi.org/10.1016/j.molstruc.2017.01.061 [ Links ]
36. Adeleke AA, Zamisa SJ, Islam MdS, Olofinsan K, Salau VF, Mocktar C, Omondi B. A study of structure-activity relationship and anion-controlled quinolinyl Ag(I) complexes as antimicrobial and antioxidant agents as well as their interaction with macromolecules. Biometals. 2022;35(2):363-394. https://doi.org/10.1007/s10534-022-00377-6 [ Links ]
37. Benzie I, Strain JJ. The ferric reducing ability of plasma (frap) as a measure of "antioxidant power": The frap assay. Anal Biochem. 1996;239(1):70-76. https://doi.org/10.1006/abio.1996.0292 [ Links ]
38. Adeleke A, Zamisa SJ, Islam MdS, Olofinsan K, Salau VF, Mocktar C, Omondi B. Quinoline functionalized schiff base silver(I) complexes: Interactions with biomolecules and in vitro cytotoxicity, antioxidant and antimicrobial activities. Molecules. 2021;26(5):1205. https://www.mdpi.com/1420-3049/26/5/1205 [ Links ]
39. Abrahams S, Mosebi S, Fish MQ, Papathanasopoulos MA, Hewer R. Screening of the nih clinical collection for inhibitors of hiv-1 integrase activity. S Afr J Chem. 2018;114(3-4):1-5. https://doi.org/10.17159/SAJS.2018%2F20170324 [ Links ]
40. Rendosova M, Vargova Z, Kuchar J, Sabolova D, Levoca S, Kudlacova J, Paulikova H, Hudecova D, Helebrandtova V, Almasi M, Vilkova M, Dusek M, Bobalova D. New silver complexes with bioactive glycine and nicotinamide molecules - characterization, DNA binding, antimicrobial and anticancer evaluation. J Inorg Biochem. 2017;168:1-12. https://doi.org/10.1016/j.jinorgbio.2016.12.003 [ Links ]
41. Patel M, Parmar PA, Gandhi DS. Synthesis, characterization and DNA binding and cleavage properties of ruthenium(II) complexes with various polypyridyls. J Enzyme Inhib Med Chem. 2011;26(5):734-741. https://doi.org/10.3109/14756366.2011.570007 [ Links ]
42. Gomathi A, Vijayan P, Viswanathamurthi P, Suresh S, Nandhakumar R, Hashimoto T. Organoruthenium(II) compounds with pyridyl benzoxazole/ benzthiazole moiety: Studies on DNA/protein binding and enzyme mimetic activities. J Coord Chem. 2017;70(10):1645-1666. https://doi.org/10.1080/00958972.2017.1309649 [ Links ]
43. Anjomshoa M, Fatemi SJ, Torkzadeh-Mahani M, Hadadzadeh H. DNA-and BSA-binding studies and anticancer activity against human breast cancer cells (mcf-7) of the zinc(II) complex coordinated by 5,6-diphenyl-3-(2-pyridyl)-1,2,4-triazine. Spectrochim Acta A Mol Biomol Spectrosc. 2014;127:511-520. https://doi.org/10.1016/j.saa.2014.02.048 [ Links ]
44. Adeleke AA, Islam MS, Omondi B. Silver(I) pyridinyl complexes with benzothiazole, thiophene, and furan moieties: DNA/protein-binding, antibacterial, antioxidant, and anticancer studies. Arch Pharm (Weinheim). 2023;356:e2200308. https://doi.org/10.1002/ardp.202200308 [ Links ]
45. Hong M, Geng H, Niu M, Wang F, Li D, Liu J, Yin H. Organotin(IV) complexes derived from schiffbase N'-[(1E)-(2-hydroxy-3-methoxyphenyl) methylidene]pyridine-4-carbohydrazone: Synthesis, in vitro cytotoxicities and DNA/bsa interaction. Eur J Med Chem. 2014;86:550-561. https://doi.org/10.1016/j.ejmech.2014.08.070 [ Links ]
46. Kong L, Yu H, Zhang J, Cui W. Tetra-aqua-bis-[N,N'-bis-(pyridin-3-yl-methyl-idene)benzene-1,4-diamine]-zinc dinitrate 1.49-hydrate. Acta Crystallogr Sect E Struct Rep Online. 2011;67(Pt 12):m1752. https://doi.org/10.1107/S1600536811046915 [ Links ]
47. Lamming G, Kolokotroni J, Harrison T, Penfold TJ, Clegg W, Waddell PG, Probert MR, Houlton A. Structural diversity and argentophilic interactions in one-dimensional silver-based coordination polymers. Crystal Growth & Design. 2017;17(11):5753-5763. https://doi.org/10.1021/acs.cgd.7b00752 [ Links ]
48. Wu H-C, Thanasekaran P, Tsai C-H, Wu J-Y, Huang S-M, Wen Y-S, Lu K-L. Self-assembly, reorganization, and photophysical properties of silver(I)-schiff-base molecular rectangle and polymeric array species. Inorg Chem. 2006;45(1):295-303. https://doi.org/10.1021/ic051235k [ Links ]
49. Pucci D, Barberio G, Bellusci A, Crispini A, La Deda M, Ghedini M, Szerb EI. Induction of columnar mesomorphism in tetracoordinated ionic silver(I) complexes based on chelate 4,4'-disubstituted 2,2'-bipyridines. Eur J Inorg Chem. 2005;2005(12):2457-2463. https://doi.org/10.1002/ejic.200500002 [ Links ]
50. Li H, Liu C, Dai B, Tang X, Zhang ZJ, Xiong Z, Liu X. Synthesis, conductivity, and electromagnetic wave absorption properties of chiral poly schiff bases and their silver complexes. J Appl Polym Sci. 2015;132(36):1-8. https://doi.org/10.1002/app.42498 [ Links ]
51. Njogu E, Omondi B, Nyamori VO. Coordination polymers and discrete complexes of Ag(I)-n-(pyridylmethylene)anilines: Synthesis, crystal structures and photophysical properties. J Coord Chem. 2017;70(16):2796-2814. https://doi.org/10.1080/00958972.2017.1370088 [ Links ]
52. Russell AD. Types of antibiotics and synthetic antimicrobial agents. Hugo and Russell's Pharmaceutical Microbiology. 2004:152-186. https://doi.org/10.1002/9780470988329.ch10 [ Links ]
53. Nandhini P, Kumar P, Mickymaray S, Alothaim AS, Somasundaram J, Rajan M. Recent developments in methicillin-resistant staphylococcus aureus (MRSA) treatment: A review. Antibiotics (Basel). 2022;11(5). https://doi.org/10.3390/antibiotics11050606 [ Links ]
54. Guentzel M. Chapter 26, escherichia, klebsiella, enterobacter, serratia, citrobacter, and proteus. Medical Microbiology, 4th Edn, ed S Baron (Galveston: University of Texas, Medical Branch) Available online at: http://www.ncbinlmnihgov/books/NBK8035/ 1996. [ Links ]
55. Patel C, Bassin JP, Scott M, Flye J, Hunter AP, Martin L, Goyal M. Synthesis and antimicrobial activity of 1,2-benzothiazine derivatives. Molecules. 2016;21(7):1-16. https://doi.org/10.3390/molecules21070861 [ Links ]
56. Matos M, Vazquez-Rodriguez S, Santana L, Uriarte E, Fuentes-Edfuf C, Santos Y, Munoz-Crego A. Synthesis and structure-activity relationships of novel amino/nitro substituted 3-arylcoumarins as antibacterial agents. Molecules. 2013;18(2):1394-1404. https://doi.org/10.3390/molecules18021394 [ Links ]
57. Pasdar H, Hedayati Saghavaz B, Foroughifar N, Davallo M. Synthesis, characterization and antibacterial activity of novel 1,3-diethyl-1,3-bis(4-nitrophenyl)urea and its metal(II) complexes. Molecules. 2017;22(12):1-12. https://doi.org/10.3390/molecules22122125 [ Links ]
58. Gök Y, Sari Y, Akkoç S, Özdemir 1, Günal S. Antimicrobial studies of n-heterocyclic carbene silver complexes containing benzimidazol-2-ylidene ligand. Int J Inorg Chem. 2014;2014:1-6. https://doi.org/10.1155/2014/191054 [ Links ]
59. Jimenez J, Chakraborty I, Rojas-Andrade M, Mascharak PK. Silver complexes of ligands derived from adamantylamines: Water-soluble silver-donating compounds with antibacterial properties. J Inorg Biochem. 2017;168:13-17. https://doi.org/10.1016/j.jinorgbio.2016.12.009 [ Links ]
60. Cifuentes-Vaca O, Andrades-Lagos J, Campanini-Salinas J, Laguna A, Vásquez-Velásquez D, Concepción Gimeno M. Silver(I) and copper(I) complexes with a schiff base derived from 2-aminofluorene with promising antibacterial activity. Inorg Chim Acta. 2019;489:275-279. https://doi.org/10.1016/j.ica.2019.02.033 [ Links ]
61. Giacomelli C, Miranda FdaS, Goncalves NS, Spinelli A. Antioxidant activity of phenolic and related compounds: A density functional theory study on the o-h bond dissociation enthalpy. Redox Rep. 2004;9(5):263-269. https://doi.org/10.1179/135100004225006038 [ Links ]
62. Irshad M, Zafaryab M, Singh M, Rizvi MM. Comparative analysis of the antioxidant activity of cassia fistula extracts. Int J Med Chem. 2012;2012:157125. https://doi.org/10.1155/2012/157125 [ Links ]
63. Adeleke AA, Islam MdS, Olofinsan K, Salau VF, Mocktar C, Omondi B. Evaluation of substituent bioactivity and anion impact of linear and t-shaped silver(I) pyridinyl complexes as potential antiproliferative, antioxidant, antimicrobial agents and DNA- and bsa-binders. New J Chem. 2021;45(38):17827-17846. https://doi.org/10.1039/d1nj03231c [ Links ]
64. El Jemli M, Kamal R, Marmouzi I, Zerrouki A, Cherrah Y, Alaoui K. Radical-scavenging activity and ferric reducing ability ofjuniperus thurifera (L.), J. Oxycedrus (L.), J. Phoenicea (L.) and tetraclinis articulata (L.). Adv Pharmacol Sci. 2016;2016:6392656. https://doi.org/10.1155/2016/6392656 [ Links ]
65. Sreelatha T, Kandhasamy S, Dinesh R, Shruthy S, Shweta S, Mukesh D, Karunagaran D, Balaji R, Mathivanan N, Perumal PT. Synthesis and sar study of novel anticancer and antimicrobial naphthoquinone amide derivatives. Bioorg Med Chem Lett. 2014;24(15):3647-3651. https://doi.org/10.1016/j.bmcl.2014.04.080 [ Links ]
66. Duric S, Vojnovic S, Andrejevic TP, Stevanovic NL, Savic ND, Nikodinovic-Runic J, Glisic BD, Djuran MI. Antimicrobial activity and DNA/bsa binding affinity of polynuclear silver(I) complexes with 1,2-bis(4-pyridyl)ethane/ ethene as bridging ligands. Bioinorg Chem Appl. 2020;2020(3812050):1-12. https://doi.org/10.1155/2020/3812050 [ Links ]
67. Banti C, Giannoulis AD, Kourkoumelis N, Owczarzak AM, Poyraz M, Kubicki M, Charalabopoulos K, Hadjikakou SK. Mixed ligand-silver(I) complexes with anti-inflammatory agents which can bind to lipoxygenase and calf-thymus DNA, modulating their function and inducing apoptosis. Metallomics. 2012;4(6):545-560. https://doi.org/10.1039/c2mt20039b [ Links ]
68. Kazemi Z, Rudbari HA, Mirkhani V, Sahihi M, Moghadam M, Tangestaninejad S, Mohammadpoor-Baltork I. Synthesis, characterization, crystal structure, DNA- and hsa-binding studies of a dinuclear schiff base Zn(II) complex derived from 2-hydroxynaphtaldehyde and 2-picolylamine. J Mol Struct. 2015;1096:110-120. https://doi.org/10.1016/j.molstruc.2015.04.033 [ Links ]
69. Saeidifar M, Jam ZS, Shahraki S, Khanlarkhani A, Javaheri M, Divsalar A, Mansouri-Torshizi H, Akbar Saboury A. Multi-spectroscopic and electrochemical approaches of the interaction between a new binuclear agent and DNA. J Biomol Struct Dyn. 2017;35(12):2557-2564. https://doi.org/10.1080/07391102.2016.1229635 [ Links ]
70. Topala T, Bodoki A, Oprean L, Oprean R. Bovine serum albumin interactions with metal complexes. Clujul Med. 2014;87(4):215-219. https://doi.org/10.15386/cjmed-357 [ Links ]
71. Lakowicz J. 1983. Protein fluorescence. Principles of fluorescence spectroscopy. Springer. p. 341-381. [ Links ]
72. Adeleke AA, Islam MdS, Sanni O, Mocktar C, Zamisa SJ, Omondi B. Aryl variation and anion effect on ct-DNA binding and in vitro biological studies of pyridinyl Ag(I) complexes. J Inorg Biochem. 2021;214:1-17. https://doi.org/10.1016/j.jinorgbio.2020.111266 [ Links ]
73. Prashanth M, Madaiah M, Revanasiddappa HD, Amruthesh KN. Synthesis, characterization, and bsa binding studies of some new benzamides related to schiff base. ISRN Org Chem. 2013;2013:791591. https://doi.org/10.1155/2013/791591 [ Links ]
Received 16 June 2022
Revised 11 March 2023
Accepted 23 March 2023
* To whom correspondence should be addressed Email: owaga@ukzn.ac.za














