Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Chemistry
On-line version ISSN 1996-840X
Print version ISSN 0379-4350
S.Afr.j.chem. (Online) vol.73 Durban 2020
http://dx.doi.org/10.17159/0379-4350/2020/v73a7
RESEARCH ARTICLE
A Spectroscopic, Thermodynamic and Molecular Docking Study of the Binding Mechanism of Dapoxetine with Calf Thymus DNA
Nawaf A. AlsaifI; Abdurrahman A. Al-MehiziaI; Ahmed H. BakheitI, III; Seema ZargarII; Tanveer A. WaniI, *
IDepartment of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, P.O. Box 2457, Riyadh 11451, Saudi Arabia
IIDepartment of Biochemistry, College of Science, King Saud University, PO Box 22452, Riyadh 11451, Saudi Arabia
IIIDepartment of Chemistry, Faculty of Science and Technology, Al-Neelain University, Khartoum, Sudan
ABSTRACT
Dapoxetine is a selective serotonin reuptake inhibitor, used to treat premature ejaculation in men. Dapoxetine may interact with the DNA and hence this study investigated dapoxetine and calf thymus DNA (ctDNA) binding interaction. The interaction study of ligands to DNA is of importance in the development of molecular probes and therapeutic agents. Spectroscopic techniques including spectrofluorometry and spectrophotometry were employed to study this interaction. Fluorescence studies indicated a static quenching mechanism between dapoxetine and ctDNA. Groove binding was suggested as the mode of interaction between dapoxetine and ctDNA based on UV absorption, circular dichroism (CD) spectroscopy, iodide quenching and molecular docking studies. The studies conducted at three different temperatures 298,303 and 310 K indicated a strong binding interaction at higher temperatures. Thermodynamic studies conducted indicated involvement of hydrophobic interaction between ctDNA and dapoxetine and were entropy-driven. Ethidium bromide probe study suggested that dapoxetine does not bind to ctDNA in an intercalative fashion. Iodide quenching studies further proved the non-intercalative binding of ctDNA with dapoxetine. Ionic strength studies conducted ruled out the electrostatic binding mechanism between ctDNA and dapoxetine. Molecular docking analysis performed for the dapoxetine with calf thymus DNA (ctDNA) interaction and confirmed minor groove binding of dapoxetine to ctDNA. The study helped to reveal the binding interaction mechanism between dapoxetine and ctDNA.
Keywords: Binding, ctDNA, dapoxetine, fluorescence, quenching.
1. Introduction
Deoxyribose nucleic acid (DNA) controls the genetic information for cell growth, division and several of its other vital functions.1 The interaction study between drugs and DNA is an important area of research because several water-soluble drugs are in their basic form and can have protonable basic groups, which are in contact with DNA molecules that are acidic.2 In the biological processes like gene expression, transcription, recombination, mutagenesis and translation, DNA plays a very essential role.
The elucidation of the binding interaction between small drug molecules and DNA has helped to understand the binding mode of a drug to DNA.2-4 Thus, such DNA-ligand binding studies are essential in the development of molecular probes and therapeutic agents. This property is used to design and synthesize new therapeutic agents.5,6 A possibility of interference in the DNA replication and RNA transcription by binding of small drug molecules to DNA may affect the gene expression.7 The two types of interaction possible between the DNA and drug molecules are covalent and non-covalent interactions. The non-covalent interactions that are possible in the drug-DNA interaction include: a) electrostatic interactions, b) groove binding and c) intercalative binding. A drug molecule binding to DNA may influence the therapeutic potential of the drug candidate and its mechanism of action.8,9
Advancement in research and availability of three dimensional DNA structure has led DNA to be an essential therapeutic drug target. Despite the progress made in DNA research, a limited number of DNA-targeted drugs have been synthesized compared to protein targeted drugs.10
Dapoxetine (+)-(S)-N,N-dimethyl-(a)-[2(1naphthalenyloxy) ethyl]- benzenemethanamine hydrochloride), a selective serotonin reuptake inhibitor, is used in the treatment of premature ejaculation. Dapoxetine is structurally similar to fluoxetine. It is absorbed rapidly and is excreted through urine with minimal accumulation in the body.11,12
The dapoxetine and ctDNA interaction was investigated with fluorescence spectroscopy, circular dichroism spectroscopy, Fourier-transform infrared spectroscopy and UV absorption spectroscopy.13,14 The principle of fluorescence quenching of dapoxetine by ctDNA was explored for this investigation to establish the mode of binding of dapoxetine to ctDNA. The thermodynamics study at three different temperatures established the binding forces involved between them. The binding site and the binding constants were also calculated in this study. Molecular docking studies helped to verify the experimental results and mode of binding between dapoxetine and ctDNA.
2. Materials and Methods
2.1. Materials
The materials were obtained from the following sources: Calf thymus DNA (ctDNA), Dapoxetine and Ethidium bromide (EB) were procured from Sigma Aldrich in the USA. Chemicals of AR grade were utilized to carry out the experiments.
2.2. Sample Preparation
Ethanol was used to prepare the stock solution of 10 mM of dapoxetine. Tris-hydrochloride buffer (pH 7.2) was used to prepare the ctDNA solution. The ctDNA was suspended in the buffer and stored at 4 °C for 12 h with occasional mixing to get a homogenous solution of ctDNA. The purity was confirmed for the ctDNA solution at A260/A280; the ratio was found to be between 1.8-1.9. The concentration of ctDNA was determined at 260 nm with a single nucleotide molar extinction coefficient of 6600 M-1 cm-1
2.3. UV-visible Spectroscopy
Shimadzu 1800 spectrophotometer, (USA), was utilized to record the UV-visible spectra. The absorption spectra for dapoxetine 0.65 μM and added ctDNA (0-50 μM) were recorded in the range of 200-400 nm.
2.4. Fluorescence Studies
The spectrofluorometer used to record the fluorescence spectra of dapoxetine was from JASCO-8200 (Japan). The instrument had a xenon flash lamp and measurements were carried out in 1.0 cm quartz cells. The λex for the spectral measurement was 287 nm and the λem were recorded in the range (300-500 nm). The slit widths used in the measurement for both excitation and emission were 5 nm. The spectra were recorded for dapoxetine (concentration was kept fixed at 0.65 μM) and ctDNA (concentration ranged between (0-50 μM)). The fluorescence studies were performed at three temperatures. The samples were kept static for 25 min at 298 K, 303 K, and 310 K before measurements.
The displacement studies with EB were conducted as follows: different concentrations of dapoxetine (0-50 /M) were added to a fixed concentration of 5 /M of EB and 50 /M of ctDNA. The spectra for EB-ctDNA were attained at λex = 524 nm and λem = 530-700 nm.
Iodide quenching experiments for dapoxetine were carried out in the presence and absence of ctDNA. Fluorescence spectra were obtained for an equimolar concentration of dapoxetine and ctDNA (50 /M) and only dapoxetine 50 /M in the presence of potassium iodide (0-8 mM).
2.5. Molecular Docking
The molecular docking study was performed on Molecular Operating Environment (MOE).15,16 The ligand structure was drawn within the MOE software. The DNA structure was downloaded from the Protein Data Bank (http://www.rcsb.org/pdb) (PDB ID: 1VTJ).17 The sequence of the downloaded DNA structure was (CGCGATATCGCG)2. The preparation of DNA for docking included geometric optimization and was performed with the MMFF94X force-field. The scoring functions 1 and 2 were London dG and GBVI/WSA dG, respectively.18
3. Result and Discussion
3.1. UV-visible Spectroscopy
The UV-visible spectroscopy being a simple technique has been exploited to investigate the interaction of ligands with proteins.19-21 Therefore, UV-visible spectroscopy was utilized to study the interaction between ctDNA and dapoxetine (Fig. 1A). Dapoxetine was observed to have three absorbance maxima at 210 nm, 230 nm and 291 nm. It has been reported that in case a ligand intercalates with the DNA helix, it leads to bathochromic shift and hypochromism.19 Hyperchromism and hypsochromic shift in absorption maxima were observed on the interaction of ctDNA with dapoxetine at 291 nm, whereas hyperchromism was observed at 210 and 230 nm (Fig. 1B). Since only hyperchromism and no bathochromic shift was observed, intercalative binding mode for ctDNA and dapoxetine was excluded.22 The change in the absorption spectrum of dapoxetine with increasing ctDNA concentration indicates complex formation between them.
3.2. Fluorescence Quenching Studies
The protein and ligand binding characteristics were studied using fluorescence quenching of fluorophores in dapoxetine by ctDNA.
The fluorescence emission spectra of dapoxetine in the presence of ctDNA (λem = 353 nm) are given in Fig. 1C. The fluorescence intensity (FI) of dapoxetine reduced in presence of increasing ctDNA concentrations.23 The λex used for the dapoxetine was 218 nm and no apparent shift in the emission wavelength was observed on its interaction with ctDNA. Thus, the quenching provides direct evidence of binding interaction between ctDNA and dapoxetine. Several molecular mechanisms are responsible for the reduced FI of certain materials and may include rearrangement of molecules; excited-state interactions; energy transfer; or complex formation. This decrease in FI is known as fluorescence quenching. The fluorescence quenching (Ksv) is determined form Stern-Volmer equation15:
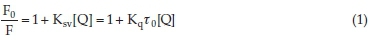
where, F0 and F represent the fluorescence intensity for dapoxetine and the dapoxetine-ctDNA system, respectively. [Q], Ksv and kq are the quencher concentration, the Stern-Volmer quenching constant and the bimolecular quenching rate constant, respectively. t0 is the fluorescence lifetime in the quencher's absence and is valued as 10-8 s.
Static and dynamic quenching are the two mechanisms involved in fluorescence quenching. An interaction between a protein and ligand can follow either a static or a dynamic quenching pattern. In static quenching, the FI of the fluorophore is quenched by the formation of a complex between the quencher and the fluorophore (non-fluorescent complex). In dynamic quenching, the FI is quenched due to collision and diffusion of molecules. Therefore, an increase in the temperature leads to higher quenching constants, since increased temperature leads to faster diffusion and high collision quenching.24
In contrast to dynamic quenching, the complexes formed between the fluorophore and the quencher in static quenching dissociate at high temperatures thus leads to a decrease in the quenching constant values.25 The quenching mechanism was identified from the behaviour of the interaction between dapoxetine -ctDNA at 298 K, 303 K, and 310 K, using Stern-Volmer plots (Fig. 2A and Table 1).26 An increase in the Ksv values suggests the involvement of dynamic or collisional quenching. However, the kq (biomolecular quenching constant) values were higher than the maximum value for bimolecular quenching rate constant of 2 x 1010 M-1 s-1, that can be obtained due to diffusion, thus suggesting complex formation and static quenching between dapoxetine and ctDNA.27,28 The increase in the absorbance of dapoxetine in presence of ctDNA, also infers complex formation and static quenching of fluorescence (Fig. 1C). In case, dynamic quenching would have been involved between dapoxetine and ctDNA, no change in the absorption intensity would have occurred, as the excited state of quencher only would have been involved.20,29

3.3. Composition of the Binary Complex
The binding interaction between the small molecules, such as drugs, with DNA, may follow one of these three binding mechanisms: intercalative binding, electrostatic binding, and groove binding. In the electrostatic binding mechanism, the cations interact with the phosphate backbone (negatively charged) of ctDNA. However, the electrostatic binding has low selectivity. The intercalation binding was first suggested by Lerman in 1961.30 In the intercalative binding, the ligand binds to the two adjacent base pairs within the DNA helix. In case of groove binding, hydrogen bonds or Van der Waals forces are formed generally in the two adjacent ctDNA double helix grooves. Further, if the binding follows static mechanism the binding equation is given as31:
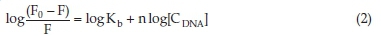
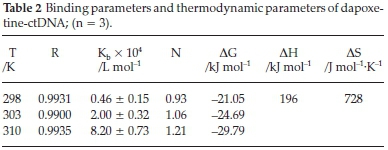
As shown in Fig. 2B, the plot of log[(F0 - F)/F] vs. log [CDNA] results in a straight line. The binding constant Kb and binding stoichiometry (n) are obtained from the slope and the intercept, respectively. The binding constants for the dapoxetine-ctDNA system were obtained at three different temperatures and are given in Table 2. The binding constants showed an increase with a rise in temperature and the value of 'n' approximated unity suggesting a single e class of binding site for dapoxetine to bind the ctDNA.16,32
3.4. Analysis of Data and Obtaining Thermodynamic Parameters
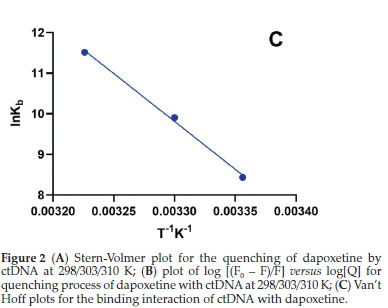
The thermodynamic parameters of entropy and enthalpy for the dapoxetine-ctDNA system were obtained from the van't Hoff plot (Fig. 2C) and equation:

The Gibb's free energy (∆G°) can be obtained from the main thermodynamic relationship equation33,34:

In the above equations, ∆H° and ∆S° represent the change in the enthalpy and entropy. Molar gas constant and temperature are represented by R and T, respectively. A straight line obtained from the plot between lnKbvs. 1/T (Fig. 2C) depicts the thermo-dynamic interaction of ctDNA with dapoxetine. The slope and the intercept of the plot are used to calculate the enthalpy and entropy, respectively. Thus, positive values obtained for ∆H° and ∆S° suggest that the dapoxetine and ctDNA interaction is entropy-driven. Further, the positive ∆H° and ∆S° indicate hydrophobic interaction between dapoxetine and ctDNA.
3.5. Circular Dichroism (CD) Spectra Studies
The conformational changes in the DNA on interaction with a ligand can be accessed by CD spectroscopic studies. In B-DNA, two CD bands are observed within the UV region. Among the two bands, the negative band appears at 245 nm and this band is due to right-handed helicity. The positive band that appears at 275 nm is due to base stalking.35 The CD spectra for ctDNA (50 μM) and increasing dapoxetine concentrations (0, 20 and 50 μM) are given in Fig. 3.

If groove binding or electrostatic force is involved in the DNA ligand interaction, the base stacking and helicity bands do not show any perturbation. In case of intercalative binding mode changes in the intensity of both the base stacking and helicity bands occur, thus stabilizing B-DNA conformation.35
No significant change in the CD spectrum was observed on addition of dapoxetine to ctDNA. No shift in the intensities of both the positive and the negative bands was observed, suggesting groove binding as a possible mode of interaction between ctDNA and dapoxetine. Further, the change in the signal of positive and negative bands on the addition of dapoxetine may be attributed to the possible transition of nucleic acid structure to a compact p structure from an extended conformation.36
3.6. FT-IR Analysis
This technique helps to characterize the ligand-ctDNA interaction and the effect of ligand over the structure of DNA. The spectral region 1000-1800 cm-1 consists most of the IR peaks of DNA. The IR peaks for ctDNA and ctDNA-dapoxetine are presented in Fig. 4. The nitorogenous bases guanine (G), thymine (T), adenine (A) and cytosine (C) of ctDNA correspond to the vibrational bands of 1714,1663,1632 and 1484 cm-1, respectively. The changes in the position of the peak of FT-IR spectra is an indicator of the ligand-DNA interaction. In the FT-IR spectra, major shifts were seen in the T band from 1663 to 1665 cm-1 and the A band from 1632 to 1626 cm-1. An increase in the intensity for all nitrogenous bases was observed. Shifts were only observed in the T and the A bands, inferring that dapoxetine selectively interacts with T and A bases. However, some interaction with G and C bases cannot be ruled out. These results, in accordance with the molecular docking results, suggest minor groove binding between ctDNA and dapoxetine.

3.7. Competitive Displacement Assay
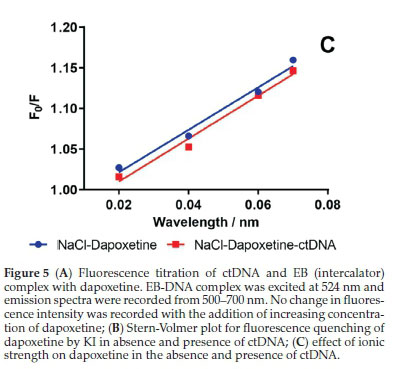
The modes of binding of dyes are well established and help in the prediction of the binding sites of drug molecules on DNA, based on displacement studies of the dye from DNA helix. The replacement of dye form the DNA helix is based on the principle that the drug is expected to bind at the same position where the dye was bound.37,38 Therefore, the FI dye-ctDNA system is expected to have some change on the addition of drug that is supposed to bind similarly. Ethidium bromide is a well-known probe dye used in the investigation of intercalative binding analysis. Hence, EB was used as a probe to confirm the binding mode between dapoxetine and ctDNA. The FI of EB-ctDNA system did not change significantly upon addition of dapoxetine (Fig. 5A). The results suggest that EB in the EB-ctDNA system is not replaced by dapoxetine and hence non-intercalative binding occurs between dapoxetine and ctDNA.
3.8. Iodide Quenching Studies
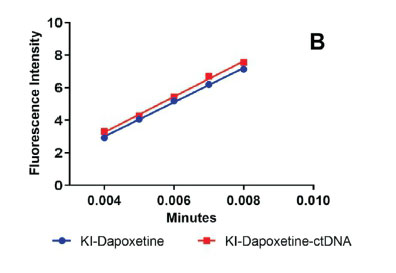
Fluorescence quenching using anionic quencher to help to deduce the binding mechanism between drug and DNA.6 In the DNA binding studies potassium iodide is used as an anionic quencher, owing to its high negative charge. In aqueous medium, iodide ions carry a negative charge and thus quench the fluorescence of drug molecules. During the interaction of iodide ions with the drug in the presence of DNA, the negative charge present on the phosphates of DNA backbone repels the negatively charged iodide ions. In case a drug molecule is intercalated within the DNA helix, it limits the access of the anionic quencher to the bound drug molecule. Hence, the drug molecule should have a larger quenching constant compared to the drug-DNA complex. Similarly, in case of groove and electrostatic binding, the access of anionic quenchers to the drug molecule is unrestricted and hence there should be no significant change in the quenching constant of free drugs and drug DNA complex.6 The Stern-Volmer constant (Ksv) is used to study the effect of anionic quencher over the drug-DNA complex. The effect on FI on dapoxetine by potassium iodide as an anionic quencher in presence of ctDNA was evaluated (Fig. 5B). No significant change was observed in the quenching constant of dapoxetine in the presence or absence of ctDNA, which were found to be 1.09 x 103 and 1.00 x 103 L mol-1, respectively. Thus, it was inferred that groove binding played a major role in binding dapoxetine to ctDNA.39
3.9. Effect of Ionic Strength
The ionic strength studies were used to identify the binding mode between small drug molecules and DNA. Sodium chloride (NaCl) is generally used only when its addition has minimal effect on the fluorescence intensity of the small drug molecule. The negative charge of the DNA phosphate backbone is neutralized by the sodium (Na+), thus, decreasing the electrostatic repulsion amid the weakening of surface-binding interactions. Also, the addition of Na+ leads to decreased electrostatic attraction amid the DNA surface and the small molecule. In electrostatic binding, the binding occurs outside the groove and addition of NaCl results in reduced quenching of FI.40
Different concentration of NaCl was used to study the effect of ionic strength on the fluorescence quenching of dapoxetine in the presence and absence of ctDNA. As is evident from Fig. 5C, an increased NaCl concentration did not change the slope of F0/F for dapoxetine in the presence or absence of ctDNA. These results indicate that the dapoxetine does not bind to ctDNA by an electrostatic binding mechanism. Also, the decrease in the FI of dapoxetine was due to the interaction of the salt with dapoxetine.
3.10. Molecular Docking
Molecular modelling of small molecules and DNA provide added information and understanding about the binding inter-action.41-44 The docking of dapoxetine with ctDNA was carried out using MOE (Fig. 6A). The docking results are in agreement with experimental results, wherein non-intercalative binding was suggested for dapoxetine and ctDNA interaction. Dapoxetine was seen to bind to the double helix ctDNA in the narrow minor groove. The minor groove binding is differentiated from its selectivity to AT base pairs.45
A weak electrostatic hydrogen bond was formed between the hydrogen atom of 5-N of dapoxetine and C4' of DT20 on chain B (Fig. 6B). The most energetically favourable binding energy found with docking study was -6.93 kcal mol-1, and it was almost similar to the binding energy -7.11 kcal mol-1 obtained experimentally.
4. Conclusions
Non-intercalative groove binding occurred between dapo-xetine and ctDNA. Complex formation between dapoxetine and ctDNA was confirmed by UV-absorption and fluorescence spectroscopic experiments. Thermodynamic parameter values suggested electrostatic and hydrophobic interaction between dapoxetine and ctDNA. Molecular docking study results were in concurrence with the experimental results, thus confirming the minor groove binding between dapoxetine and ctDNA.
Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research, King Saud University, for funding the research group No. RG-1473-073.
Conflicts of Interest
The authors have declared no conflict of interest.
ORCID iDs
A.A. Al-Mehizia: orcid.org/0000-0001-8711-3873
S.Zargar: orcid.org/0000-0002-5622-0841
References
1 B. Lewin and G. Dover, Genes v. Vol. 1110, Oxford University Press, Oxford, 1994. [ Links ]
2 L. Alarcón, Y. Baena and R. Manzo, Interaction between DNA and drugs having protonable basic groups: characterization through affinity constants, drug release kinetics, and conformational changes, Scient. Pharmaceut., 2017, 85, 1. [ Links ]
3 A. Chandran, D. Ghoshdastidar and S. Senapati, Groove binding mechanism of ionic liquids: a key factor in long-term stability of DNA in hydrated ionic liquids?, J. Am. Chem. Soc, 2012,134, 20330-20339. [ Links ]
4 N. Shahabadi and L. Heidari, Binding studies of the antidiabetic drug, metformin to calf thymus DNA using multispectroscopic methods, Spectrochim. Acta Part A: Mol. Biomol. Spectrosc., 2012, 97, 406-410. [ Links ]
5 F. Cui, G. Hui, X. Jiang and G. Zhang, Interaction of 3'-azido-3'-diamino daunorubicin with DNA: multispectroscopic and molecular modeling, Int. J. Biol. Macromol, 2012, 50, 1121-1126. [ Links ]
6 S.U. Rehman, Z. Yaseen, M.A. Husain, T Sarwar, H.M. Ishqi and M. Tabish, Interaction of 6 mercaptopurine with calf thymus DNA-deciphering the binding mode and photoinduced DNA damage, PLOS ONE, 2014, 9, e93913. [ Links ]
7 J.B. Chaires, Drug-DNA interactions, Curr. Opin. Struct. Biol., 1998,8, 314-320. [ Links ]
8 A.Y. Chen and L.F. Liu, DNA topoisomerases: essential enzymes and lethal targets, Annu. Rev. Pharmacol. Toxicol., 1994, 34, 191-218. [ Links ]
9 D.S. Pilch, M.A. Kirolos, X. Liu, G.E. Plum and K.J. Breslauer, Berenil [1, 3-bis (4'-amidinophenyl) triazene] binding to DNA duplexes and to a RNA duplex: evidence for both intercalative and minor groove binding properties, Biochem. (Mosc.). 1995, 34, 9962-9976. [ Links ]
10 H. Berman, J. Westbrook, Z. Feng, etal., The protein databank. Nucleic Acid Res, 2000, 28, 235-242. [ Links ]
11 E. McCarty and W Dinsmore, Dapoxetine: an evidence-based review of its effectiveness in treatment of premature ejaculation, Core Evidence, 2012, 7, 1. [ Links ]
12 C.G. McMahon, Dapoxetine: a new option in the medical management of premature ejaculation, Therap. Adv. Urol., 2012, 4, 233-251. [ Links ]
13 S. Khatun, Probing of the binding profile of anti-hypertensive drug, captopril with bovine serum albumin: a detailed calorimetric, spectroscopic and molecular docking studies, J. Chem. Thermodynam., 2018,126, 43-53. [ Links ]
14 X. Wang and F. Cui, Binding characteristics of imidazolium-based ionic liquids with calf thymus DNA: spectroscopy studies, J. Fluorine Chem., 2018, 213, 68-73. [ Links ]
15 T.A. Wani, A.H. Bakheit, M. Abounassif and S. Zargar, Study of interactions of an anticancer drug neratinib with bovine serum albumin: spectroscopic and molecular docking approach, Frontiers Chem., 2018, 6, 47. [ Links ]
16 T.A. Wani, A.H. Bakheit, M.N. Ansari, A.-R.A. Al-Majed, B.M. Al-Qahtani and S. Zargar, Spectroscopic and molecular modeling studies of binding interaction between bovine serum albumin and roflumilast, Drug Des., Dev. Therap, 2018,12, 2627. [ Links ]
17 PW Rose, A. Prlic, A. Altunkaya, et al., The RCSB protein data bank: integrative view of protein, gene and 3D structural information, Nucleic Acids Res., 2016, gkw1000. [ Links ]
18 T.A. Wani, A.H. Bakheit, S. Zargar, M.A. Bhat and A.A. Al-Majed, Molecular docking and experimental investigation of new indole derivative cyclooxygenase inhibitor to probe its binding mechanism with bovine serum albumin, Bioorg. Chem., 2019, 89, 103010. [ Links ]
19 A. Pyle, J. Rehmann, R. Meshoyrer, C. Kumar, N. Turro and J.K. Barton, Mixed-ligand complexes of ruthenium (II): factors governing binding to DNA, J. Am. Chem. Soc., 1989, 111, 3051-3058. [ Links ]
20 T.A. Wani, A.H. Bakheit, A.-R.A. Al-Majed, M.A. Bhat and S. Zargar, Study of the interactions of bovine serum albumin with the new anti-inflammatory agent 4-(1, 3-dioxo-1, 3-dihydro-2H-isoindol-2-yl)-N'-[(4-ethoxy-phenyl) methylidene] benzohydrazide using a multi-spectroscopic approach and molecular docking, Molecules, 2017, 22, 1258. [ Links ]
21 T.A. Wani, A.H. Bakheit, S. Zargar, M.A. Hamidaddin and I.A. Darwish, Spectrophotometric and molecular modelling studies on in vitro interaction of tyrosine kinase inhibitor linifanib with bovine serum albumin, PLOS ONE, 2017,12, e0176015. [ Links ]
22 S.M. de Almeida, E.A. Lafayette, L.P. da Silva, et al., Synthesis, DNA binding, and antiproliferative activity of novel acridine-thiosemi-carbazone derivatives, Int. J. Mol. Sci., 2015,16, 13023-13042. [ Links ]
23 G. Rabbani, E.J. Lee, K. Ahmad, M.H. Baig and I. Choi, Binding of tolperisone hydrochloride with human serum albumin: effects on the conformation, thermodynamics, and activity of HSA, Mol. Pharm., 2018,15, 1445-1456. [ Links ]
24 G. Rabbani, M.H. Baig, A.T. Jan, et al., Binding of erucic acid with human serum albumin using a spectroscopic and molecular docking study, Int. J. Biol. Macromol., 2017,105, 1572-1580. [ Links ]
25 G. Rabbani, M.J. Khan, A. Ahmad, M.Y. Maskat and R.H. Khan, Effect of copper oxide nanoparticles on the conformation and activity of ß-galactosidase, Colloids Surf., B. Biointerf., 2014,123, 96-105. [ Links ]
26 S.M. Abdullah, S. Fatma, G. Rabbani and J.M. Ashraf, A spectroscopic and molecular docking approach on the binding of tinzaparin sodium with human serum albumin, J. Mol. Struct., 2017,1127,283-288. [ Links ]
27 G. Rabbani, M.H. Baig, E.J. Lee, W.-K. Cho, J.Y. Ma and I. Choi, Biophysical study on the interaction between eperisone hydrochloride and human serum albumin using spectroscopic, calorimetric, and molecular docking analyses, Mol. Pharm., 2017,14, 1656-1665. [ Links ]
28 G. Rabbani, E. Ahmad, N. Zaidi and R.H. Khan, pH-dependent conformational transitions in conalbumin (ovotransferrin), a metallo-proteinase from hen egg white, Cell Biochem. Biophys., 2011,61,551-560. [ Links ]
29 N. Shahabadi, S. Hadidi and F. Feizi, Study on the interaction of antiviral drug 'Tenofovir' with human serum albumin by spectral and molecular modeling methods, Spectrochim. Acta Part A: Mol. Biomol. Spectrosc, 2015,138, 169-175. [ Links ]
30 L. Lerman, Structural considerations in the interaction of DNA and acridines, J. Mol. Biol., 1961, 3, 18-IN14. [ Links ]
31 A.H. Hegde, S. Prashanth and J. Seetharamappa, Interaction of anti-oxidant flavonoids with calf thymus DNA analyzed by spectroscopic and electrochemical methods, J. Pharm. Biomed. Anal., 2012,63,40-46. [ Links ]
32 S. Bakkialakshmi and D. Chandrakala, Fluorometric investigation on the interaction of 5-bromo and 5-iodouracil with bovine serum albumin, Archiv. Appl. Sci. Res., 2011, 3, 174-179. [ Links ]
33 A.M. Alanazi, A.S. Abdelhameed, A.H. Bakheit, et al., Spectroscopic and molecular docking studies of the binding of the angiotensin II receptor blockers (ARBs) azilsartan, eprosartan and olmesartan to bovine serum albumin, J. Lumin., 2018, 203, 616-628. [ Links ]
34 M.M. Alanazi, A.A. Almehizia, A.H. Bakheit, N.A. Alsaif, H.M. Alkahtani and T.A. Wani, Mechanistic interaction study of 5, 6-dichloro-2-[2-(pyridin-2-yl) ethyl] isoindoline-1,3-dione withbovine serum albumin by spectroscopic and molecular docking approaches, Saudi Pharm. J., 2018. [ Links ]
35 X. Zhou, G. Zhang and L. Wang, Probing the binding mode of psoralen to calf thymus DNA, Int. J. Biol. Macromol., 2014, 67, 228237. [ Links ]
36 T. Sarwar, H.M. Ishqi, S.U. Rehman, M.A. Husain, Y. Rahman and M. Tabish, Caffeic acid binds to the minor groove of calf thymus DNA: a multi-spectroscopic, thermodynamics and molecular modelling study, Int. J. Biol. Macromol., 2017, 98, 319-328. [ Links ]
37 Z. Mirzaei-Kalar, In vitro binding interaction of atorvastatin with calf thymus DNA: multispectroscopic, gel electrophoresis and molecular docking studies, J. Pharm. Biomed. Anal., 2018, 161, 101-109. [ Links ]
38 Y. LV P. Li, M.-L. Jiao, B.-S. Liu and C. Yang, Fluorescence quenching study of moxifloxacin interaction with calf thymus DNA, Turkish J. Chem., 2014, 38, 202-209. [ Links ]
39 D. Sarkar, P. Das, S. Basak and N. Chattopadhyay, Binding interaction of cationic phenazinium dyes with calf thymus DNA: a comparative study, J. Phys. Chem. B, 2008, 112, 9243-9249. [ Links ]
40 M. Tao, G. Zhang, C. Xiong and J. Pan, Characterization of the interaction between resmethrin and calf thymus DNA in vitro, New J. Chem, 2015, 39, 3665-3674. [ Links ]
41 X. Zhou, G. Zhang and J. Pan, Groove binding interaction between daphnetin and calf thymus DNA, Int. J. Biol. Macromol., 2015, 74, 185-194. [ Links ]
42 A.M. Alanazi, A.S. Abdelhameed, A.H. Bakheit, et al., Unraveling the binding characteristics of the anti-HIV agents abacavir, efavirenz and emtricitabine to bovine serum albumin using spectroscopic and molecular simulation approaches, J. Mol. Liq., 2018, 251, 345-357. [ Links ]
43 E. Ahmad, G. Rabbani, N. Zaidi, et al., Stereo-selectivity of human serum albumin to enantiomeric and isoelectronic pollutants dissected by spectroscopy, calorimetry and bioinformatics, PLOS ONE, 2011, 6, e26186. [ Links ]
44 A. Varshney, M. Rehan, N. Subbarao, G. Rabbani and R.H. Khan, Elimination of endogenous toxin, creatinine from blood plasma depends on albumin conformation: site specific uremic toxicity & impaired drug binding, PLOS ONE, 2011, 6, e17230. [ Links ]
45 W. Müller and D.M. Crothers, Interactions of heteroaromatic compounds with nucleic acids: 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands, Eur. J. Biochem., 1975, 54, 267-277. [ Links ]
Received 14 October 2019
Revised 16 February 2020
Accepted 17 February 2020
* To whom correspondence should be addressed. E-mail: twani@ksu.edu.sa