Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Chemistry
On-line version ISSN 1996-840X
Print version ISSN 0379-4350
S.Afr.j.chem. (Online) vol.66 Durban Aug. 2013
RESEARCH ARTICLE
Synthesis, characterization and antibacterial activity of imidazole derivatives of 1,10-phenanthroline and their Cu(II), Co(II) and Ni(II) complexes
Mesut GomleksizI, *; Cihan AlkanII; Belgin ErdemIII
IDepartment of Chemistry, Faculty of Science and Arts, Ahi Evran University, TR-40100, Kirsehir, Turkey
IIDepartment of Chemistry, Faculty of Science and Arts, Firat University, TR-23119, Elazig, Turkey
IIIDepartment of Biology, Faculty of Science and Arts, Ahi Evran University, TR-40100, Kirsehir, Turkey
ABSTRACT
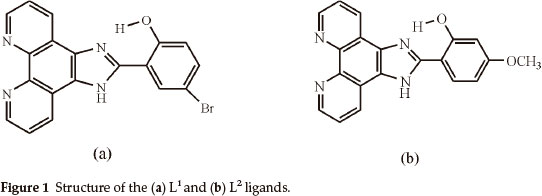
Six new CuL1 (L1 = 4-bromo-2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)phenol), CoL1, NiL1, CuL2 (L2 = 2-(1H-imidazo[4,5-f] [1,10]phenanthroline-2-yl)-5-methoxyphenol), CoL2 and NiL2 complexes were synthesized. L1 and L2 ligands were prepared by the condensation of 1,10-phenanthroline-5,6-dione with 5-bromosalicylaldehyde and 2-hydroxy-4-methoxybenzaldehyde, respectively. The structures of the compounds were determined by elemental analyses, IR, UV-visible, 1H-NMR, TGA, magnetic susceptibilities and molar conductance measurements. It is observed that the synthesized complexes have tetragonal and distorted square pyramidal geometrical structures. Antibacterial activity of the ligands and their metal complexes were tested against selected bacteria by disc diffusion method.
Keywords: 1,10-Phenanthroline, imidazole, complex, antibacterial activity.
1. Introduction
1,10-Phenanthroline (phen) and its derivations play important roles for supramolecular assemblies because they can also provide bidentate N-donor sites for chelating with metal ions to form bridge ligands.1-4 Derivatives of phen are very important ligands in organometallic chemistry;5,6 some of their complexes, for example, bind to DNA.7-10
Metal complexes of the type [M(LL)3]n+ where LL is either phen or a modified phen ligand, are particularly attractive species to recognize and cleave DNA.11-13 Systematic studies of substituted derivatives of phen have been successfully undertaken.14 1,10-phenanthroline, as well as some of its derived complexes, do exhibit antimicrobial properties.15,16 The photocemical and redox properties of complexes can be varied systematically through appropriate substition on the phenanthroline rings.17,18
Firstly, we synthesized and characterized Cu(II), Co(II) and Ni(II) complexes with phen imidazole derivatives, which are 4-bromo-2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)phenol (L1) and 2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)-5-methoxyphenol (L2) (Fig. 1). Secondly, these compounds were screened for antibacterial activity against such bacterial strains as A. hydrophila, S. aureus, K. pneumoniae, P. aeruginosa, S. marces-cens, E. aerogenes, B. subtilis, E. coli and E. faecalis.
2. Experimental
2.1. Materials and Physical Measurements
1,10-phenanthroline-5,6-dione was synthesized according to a published method.19 Ethanol was dried over anhydrous copper (II) sulfate and distilled over metallic sodium. All other chemicals were of analytical grade and were used as purchased.
Elemental analyses (C, H, N) were performed by using a Leco 932 elemental analyzer. 1H NMR spectra were recorded on a Bruker 300 MHz spectrometer in DMSO-d6. The IR spectra were obtained using KBr discs on an Ati Unicam Mattson 1000 Series FT-IR spectrophotometer. The electronic absorption spectra in the 200-1100 nm range were obtained in DMF on a Shimadzu UV-1700 UV-Visible spectrophotometer. Magnetic susceptibility measurements were carried out by the Gouy method at room temperature using Hg[Co(SCN)4] as a reference for calibrant. Conductivities of a 10-3 M solution of the complexes were measured in DMF at 25 °C using a CMD 750 WPA model conductivity meter. Thermogravimetric analyses (TGA) were carried out by Shimadzu-50 thermal analyzer in a dynamic nitrogen atmosphere in the 20-600 °C range and a heating rate of 20 °C min-1.
2.2. Antibacterial Activity
The in vitro antibacterial screening effects of the ligands (L1,L2) and their metal complexes were tested against nine bacterial strains, namely A. hydrophila ATCC 7966, S. aureus ATCC 29213, K. pneumoniae ATCC 21541, P. aeruginosa ATCC 27853, S. marcescens ATCC 21074, E. aerogenes ATCC 5402, B. subtilis ATCC 6633, E. coli ATCC 25922 and E. faecalis ATCC 29212 by disc diffusion method using nutrient agar medium for antibacterial activity.20 All bacteria were inoculated into Nutrient Broth (Difco) and incubated for 24 h. In the agar well diffusion method (Mueller Hinton Agar (Oxoid) for bacteria), the dilution plate method was used to enumerate microorganisms (105 bacteria per mL) for 24 h.21 Using a sterilized cork borer (6 mm diameter), wells were dug in the culture plates. Metal complexes and ligands were performed at the fixed concentration of 2000 mL-1 and compounds dissolved in DMF. Compounds dissolved in DMF were added (75 µL.) to these wells. The petri dishes were left at 4 °C for 2 h and then the plates were incubated at 37 °C and 30 °C for bacteria (18-24 h). At the end of the period, inhibition zones formed on the medium were evaluated is milimetres. DMF was used as negative control under similar conditions for comparison. Ampicillin (AMP) was used as the reference drug in positive controls. The experiments were performed in triplicate.
2.3. Statistical Analysis
In this study, repeated measures analysis of variance was used to evaluate the data. Ligands and their metal complexes were analyzed antibacterial activity at different temperatures. Statistical significance was determined using Duncan multiple comparison test and Bonferroni multiple comparison test was used for grouping within subject factors. SPSS 15.0, version 8, software was used in the statistical analyses.22
2.4. Synthesis of Ligands (L1, L2)
Ligands (L1, L2) were synthesized by a method similar to one described previously.18
4-bromo-2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)phenol (L1)
A mixture of 1,10-phenanthroline-5,6-dione (0.2 g, 1 mmol), ammonium acetate (1.54 g, 20 mmol), 5-bromosalicylaldehyde (0.22 g, 1.1 mmol) and glacial acetic acid (25 mL) was refluxed for 2 h, then cooled to room temperature and diluted with water (50 mL). Dropwise addition of concentrated aqueaus ammonia to neutralize gave a yellow precipitate, which was collected and washed with water. The crude product dissolved in ethanol was purified by filtration on silica gel. The principal yellow band was collected. Evaporation of the solution gave yellow crystals. It was filtered, washed with ethanol and recrystallized from ethanol then dried at 80 °C. Yield: 0.311 g (79 %). IR (v,cm-1): 3245-2420 (N-H and O-H.-N), 1607 (C=N imidazole ring), 1574, 1563, 1539, 1503 (C=C aromatic and C=N phenanthroline ring); δH (300 MHz, DMSO-d6): 13.6 (1H, s, OH), 12.82 (1H, s, NH), 8.97-8.91 (2H, d, CAr - H), 8.63-8.56 (2H, dd, CAr - H), 8.18-8.13 (1H, d, CAr-H), 7.71-7.61 (2H, dd, CAr-H), 7.46-7.39 (1H, dd, CAr-H) and 6.98-6.92 ppm (1H, d, CAr - H); UV-Vis (in DMF, nm): 277, 286, 303, 324, 337, 357, 407 and 552.
2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)-5-methoxyphenol (L2)
L2 was synthesized by a procedure similar to that for L1 except that 2-hydroxy-4-methoxybenzaldehyde was used, and was obtained as yellow powder. Yield: 0.147 g (43 %). IR (v,cm-1): 3274-2456 (N-H and O-H-.N), 1604 (C=N imidazole ring), 1591, 1563, 1544, 1508 (C=C aromatic and C=N phenanthroline ring), 1256 (Ar-O-CH3); δH (300 MHz, DMSO-d6): 15.81 (1H, s, Oh), 12.85 (1H, s, NH), 9.06-8.81 (4H, m, CAr - H), 7.86-7.71 (3H, m, CAr-H), 7.11-6.97(2H,m, CAr-H) and 3.86 (3H, s, OCH3); UV-Vis (in DMF, nm): 279, 340, 356, 413, 445 and 550.
2.5. Synthesis of Complexes
CuL1 CoL1 and NiL1
A solution of a metal salt (0.1 mmol) in DMF (2 mL) was added to a hot solution of the L1 (0.078 g, 0.2 mmol) in DMF (10 mL).The reaction mixture was heated at 80 °C until the reaction was complete. The mixture was then left for two weeks at room temperature, filtered, washed with DMF, water and ethanol and dried at 100 °C in a vacuum oven. The following salts were used for the synthesis; CuCl2.H2O (0.017 g, 10 h reaction time), CoCl2.6H2O (0.030 g, 24 h reaction time), NiCl2.6H2O (0.020 g, 10 h reaction time).
CuL1: Green compound. Yield: 0.048 g (52 %). IR (v,cm-1): 3208-2540 (N-H and O-H...N),1604(C=N imidazole ring), 1580, 1541, 1511 (C=C aromatic and C=N phenanthroline ring); UV-Vis (in DMF, nm): 453 and 700; (Found: C, 48.96; H, 2.54; N, 11.97 %. Calc. for C38H22N8O2Cl2Br2Cu (916.89); C, 49.78; H, 2.42; N, 12.22 %); µeff: 1.86 BM; ΛM (10-3 M, in DMF, Ω-1 cm2 mol-1): 12.43.
CoL1: Brown compound. Yield: 0.049 g (53 %). IR (v,cm-1): 3428 (O-H, H2O), 3190-2406 (N-H and O-H...N), 1607 (C=N imidazole ring), 1583, 1541, 1511 (C=C aromatic and C=N phenanthroline ring); UV-Vis (in DMF, nm): 502; (Found: C, 50.21; H, 3.11; N, 11.50 %. Calc. for C38H24N8O3Cl2Br2Co (930.30); C, 49.06; H, 2.60; N, 12.04 %); µeff: 4.83 BM; ΛM (10-3 M, in DMF, Ω-1 cm2 mol-1): 76.84.
NiL1: Orange compound. Yield: 0.066 g (63 %). IR (v,cm-1): 3126-2460 (N-H and O-H.-N), 1604 (C=N imidazole ring), 1583, 1541, 1511 (C=C aromatic and C=N phenanthroline ring); UV-Vis (in DMF, nm): 452; (Found: C, 43.87; H, 2.88; N, 10.58 %. Calc. for C38H22N8O2Cl4Br2Ni2 (1041.64); C, 43.82; H, 2.13; N, 10.76 %); µeff: 1.53 BM; ΛM (10-3 M, in DMF, Ω-1 cm2 mol-1): 10.34.
CuL2: A ethanolic (20 mL) solution of the (0.025 g, 0.15 mmol) CuCl2.H2O was added to a hot ethanolic (40 mL) solution of the L2 (0.100 g, 0.3 mmol). The mixture was refluxed for 24 h. The mixture was cooled to room temperature, the resulting green solid was filtered, washed with DMF and ethanol then dried at 100 °C in a vacuum oven. Yield: 0.082 g (67 %). IR (v,cm-1): 3215-2405 (N-H and O-H.-N), 1604 (C=Nimidazole ring), 1591, 1577,1544,1508 (C=C aromatic and C=N phenanthroline ring), 1248 (Ar-O-CH3); UV-Vis (in DMF, nm): 470; (Found: C, 59.20; H, 4.06; N, 12.93 %. Calc. for C40H28N8O4Cl2Cu (819.15); C, 58.65; H, 3.45; N, 13.68 %); µeff: 2.13 BM; ΛM (10-3 M, in DMF, Ω-1 cm2 mol-1): 14.82.
CoL2 and NiL2
A solution of a metal salt (0.15 mmol) in DMF (2 mL) was added to a hot solution of the L2 (0.100 g, 0.3 mmol) in DMF (10 mL).The mixture was heated at 80 °C while stirring for 24 h. The mixture was left for two weeks at room temperature, the resulting solid was filtered, washed with DMF, water and ethanol then dried at 100 °C in a vacuum oven. The following salts were used for the synthesis; CoCl2.6H2O (0.038 g), NiCl2.6H2O (0.036 g).
CoL2: Brown compound. Yield: 0.057 g (46 %). IR (v,cm-1): 3423 (O-H, H2O), 3115-2400 (N-H and O-H-N), 1604 (C = N imidazole ring), 1591,1577,1544,1508 (C = C aromatic and C=N phenanthroline ring), 1248 (Ar-O-CH3); UV-Vis (in DMF, nm): 487, 606, 926; (Found: C, 58.56; H, 4.06; N, 13.15 %. Calc. for C40H30N8O5Cl2Co (832.56); C, 57.71; H, 3.63; N, 13.46 %); µeff: 4.92 BM; ΛΜ (10-3 M, in DMF, Ω-1 cm2 mol-1): 17.65. NiL2: Orange compound. Yield: 0.052 g (40 %).IR (v,cm-1): 3410 (O-H, H2O), 3115-2280 (N-H and O-H...N), 1604 (C=N imidazole ring), 1591,1577,1544,1508 (C = C aromatic and C=N phenanthroline ring), 1248 (Ar-O-CH3); UV-Vis (in DMF, nm): 500, 595, 902; (Found: C, 55.16; H, 3.98; N, 11.63 %. Calc. for C40H34N8O7CI2Ni (868.35); C, 55.33; H, 3.95; N, 12.90 %); µeff: 3.26 BM; ΛΜ (10-3 M, in DMF, Ω-1 cm2 mol-1): 148.42.
3. Results and Discussion
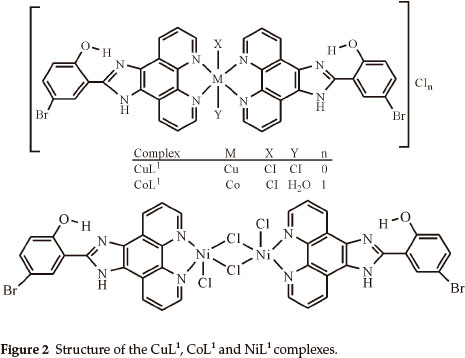
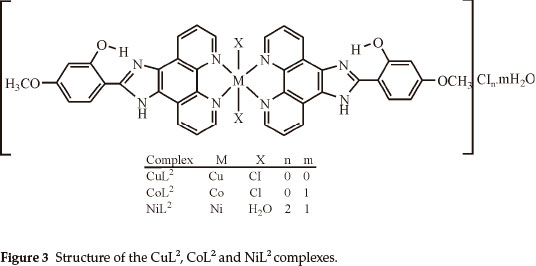
Elemental analyses indicate that the metal:ligand ratio is 1:1 in the case of the NiL1 complex and 1:2 in the case of the other complexes. In addition, the magnetic moment value of NiL1 complex indicates a dimeric structure (Figs. 2,3). The ligands L1 and L2 were soluble in EtOH, DMF and DMSO, and the complexes in DMF and DMSO. The melting points of the all compounds were not observed due to decomposition.
3.1. IR Spectra
In IR spectra of CoL1, CoL2 and NiL2, the bands are observed at the 3428, 3423 and 3410 cm-1 as broad bands are due to the OH stretching vibrations of H2O molecules.23,24,25
The broadened band between 3274-2420 cm-1 in IR spectra of the L1 and L2 ligands is due to the stretching vibrations of the both NH of the imidazole ring and intramolecular hydrogen bonding (O-H...N) formed between phenolic OH and nitrogen atom of C=N group of imidazole ring.26 The same band was observed in IR spectra of metal complexes of these ligands. This observation confirmed that phenolic OH and nitrogen (C=N) of the imidazole ring do not participate in coordination. Moreover, the stretching vibration of the C=N group (imidazole ring) of the ligands L1 and L2 were not significantly affected in their complexes, indicating that the nitrogen atom of this group is not involved in coordination for all the complexes. On the other hand, the bands of the C=N (phenanthroline ring) and C=C (Aromatic) groups were shifted to higher frequencies in all the complexes of L1 and the band at 1563 cm-1 in the free L2 ligand was shifted to higher frequencies (1577 cm-1) in their complexes,27,28 that indicates the participation of the C=N (phenanthroline ring) groups in coordination of the metal ion.
The bands of the N-H and O-H...N groups in all the complexes of L1 shifted to negative frequencies after complexations. The N-H, O-H...N and Ar-O-CH3 groups in all complexes of L2 are the some as complexes of L1. The negative frequency shifts of these groups may be attributed to flow of electrons from these groups to the phenanthroline ring due to electron flow from the nitrogen atom of the phenanthroline ring to the metal ion after complexations.
3.2. Electronic Spectra and Magnetic Measurements
In the electronic spectra of L1 and L2 ligands, the bands are observed in the range of 277-552 nm and 279-550 nm, respectively. These bands are attributed to π→π* and n→ π* transitions.29,30
The magnetic moment values for the Cu(II) complexes lies in the range 1.86-2.13 BM corressponding to one unpaired electron.31 The complexes may be considered to possess a tetragonal geometry. The electronic spectra of CuL1 complex shows two bands at 453 and 700 nm assigned to 2B1g → (2B2g, 2Eg) and 2B1g → 2A1g transitions, respectively. The band observed at 470 nm for CuL2 complex is assigned to 2Ε(D4h; 2B1g, 2A1g) → 2T2g(D4g; 2Eg, 2B2g). The band assigned to 2B1g → 2A1g transition cannot be detected for the CuL2 complex.32
The magnetic moment values for the Co(II) complexes in the range 4.83-4.92 BM reported here show that there are three unpaired electrons, indicating a high spin octahedral configura-tion.31 The electronic spectra of CoL2 complex give three bands 487, 606 and 926 nm. These bands may be assigned to the transitions 4T1g(F)(D4h; 4A2g,4Eg) → 4T1g(P)(D4h; 4A2g,4Eg)(F)(D4h; 4A2g,, 4Eg) → 4A2g(F)(D4h; 4B1g) and 4T1g(F)(D4h; 4A2g,4Eg)→4T2g(F)(D4h; 4B2g, 4Eg), respectively. The band observed at 502 nm for the CoL1 complex is assigned to 4Eg → T1g(P)(4Eg). The positions of these bands suggest a tetragonal environment around Co2+ ion.32 The other bands of CoL1 are not observed because they might be overlap with bands of the L1 ligand.
The low µeff (1.53 BM) of NiL1 complex indicate a dimeric structure.31,33 The band observed at 452 nm for NiL1 complex is assigned to 3B1 → 3A23E(P) of a distorded square pyramidal structure. The other band assigned to 3B1 → 3B2is not observed because it overlaps with ligand bands of L1. The magnetic moment value (3.26 BM) for NiL2 corresponds to two unpaired electron.31 The electronic spectrum of this complex shows absorption bands at 500,595 and 902 nm, attributed to 3A2g(D4h; 3B1g) → 3T1g(P)(D4h; 3Eg, 3A2g),3A2g(D4h;3B1g)-3T1g(F)(D4h;3Eg,3A2g) and 3A2g(D4h;3B1g)- 3T2g(F)(D4h; 3B2g, 3Eg) transitions, respectively, in a tetragonal geometry around the Ni2+ ion.32
3.3. Thermal Analysis (TGA)
According to the thermogravimetric results CuL1, NiL1, and CuL2 exhibited rather high thermal stability with decomposition temperatures of 320,280 and 270 °C, respectively. CoL1, CoL2 and NiL2 complexes were stable up to 175,50 and 50 °C, respectively. In the decomposition process of these complexes, the mass loss corresponded to one coordinated water molecule in the temperature range 175-240 °C for CoL1 (2.08 % experimental; 1.93 % calculated), one uncoordinated water molecule in the temperature range 50-100 °C for CoL2 (2.50 % experimental; 2.60 % calculated) and NiL2 (2.08 % experimental; 2.10 % calculated). In the second stage of the decomposition process of NiL2 the mass loss corresponded to two coordinated water molecules in the temperature range 160-250 °C (4.16 % experimental; 4.14 % calculated).
3.4. Conductance Measurements
Conductivity measurements of CoL1 complex resulted in ΛM 76.84 Ω-1cm2 mol-1, which indicates that it is of the 1:1 electrolyte type. NiL2 had an ΛM value of 148.42 Ω-1 cm2 mol-1) indicating that it is of the 2:1 electrolyte type. The other complexes were nonelectrolytes.34
3.5. Antibacterial Activity
Test results of antibacterial screening are summarized in Tables 1 and 2. According to the results of the antibacterial activity, ligands and all their metal complexes showed antibacterial activity at 30 °C and 37 °C in which there is no distinction between antibacterial activity. The Duncan's multiple range test indicated significant differences of antibacterial activity among ligands and their metal complexes. The ligands displayed weak antibacterial activity against B. subtilis. However, good activity was observed against others bacteria. Cu(II) complexes displayed good ant bacter al act v ty aga nst all bacter a except for B. subtilis. Co(II) complexes exhibited activity against S. aureus, B. subtilis, A. hydrophila, K. pneumoniae, E. aerogenes and E. coli. However, no activity was observed against S. marcescens, E. coli and P. aeruginosa. Additionally, Ni(II) complexes exhibited weak effect against to all bacteria tested.
Finally, these results may suggest that the ligands and their metal complexes can be used as antibacterial agents in new drugs for therapy of infectious diseases in humans.
4. Conclusion
In this study, imidazole and phenanthroline containing 4-bromo-2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)phenol (L1), 2-(1H-imidazo[4,5-f][1,10]phenanthroline-2-yl)-5-metho-xyphenol) (L2) and their complexes were synthesized and characterized. The analytical data and spectroscopic studies suggest that the complexes were of the general formula: [M(L1)2XY]CIn where M is Cu(II) (X = CI, Y = CI, n = 0), Co(II) (X = CI, Y = H2O, n = 1), [M2(L1)2CI4] where M = Ni(II) and [M(L2)2X2]CInmH2O where M is Cu(iI) (x = CI, n = 0, m=0), Co(II) (X = CI, n = 0, m=1) and Ni(II) (X = H2O, n = 2, m=1). According to the IR data of the compounds, ligands (L1,L2) are coordinated to the metal ions through nitrogen atoms of the C=N (phenanthroline ring) groups.
The results obtained from this research demonstrated that all synthesized compounds have antibacterial activity against the bacterial strains. In this sense, we think that the ligands and their metal complexes might be effective as antibacterial agents against bacteria.
Acknowledgements
We are grateful to Firat University Research Foundation for the support of this research.
References
1 S. Bodige and F.M. MacDonnell, Tetrahedron Lett., 1997, 38, 8159-8160. [ Links ]
2 W. Paw and R. Eisenberg, Inorg. Chem, 1997, 36, 2287-2293. [ Links ]
3 K. Nobuko, K. Ryuzi, H. Yuichiro, S. Hideki, A. Hironori, and K. Kazuyuki, J. Chem. Soc., Dalton Trans., 2000, 18, 3053-3054. [ Links ]
4 V Balzani, A. Juris, M. Venturi, S. Campagna and S. Serroni, Chem. Rev., 1996, 96, 759-833. [ Links ]
5 E.C. Constable, Metals and Ligands Reactivity, VCH, Weinheim, 1996, p. 308. [ Links ]
6 J.P. Sauvage, J.P. Collin, J.C. Chambron, S. Guillerez, C. Coudret, V. Balzani, F. Barigeletti, L. De Cola and L. Flamingi, Chem. Rev., 1994, 94, 993-1019. [ Links ]
7 R.E. Holmlin, P.J. Dandliker and J.K. Barton, Angew. Chem, 1997, 36, 2714-2730. [ Links ]
8 F. Gao, H. Chao, J.Q. Wang, Y.X. Yuan, B. Sun, Y.F. Wei, B. Peng and L.N. Ji, J. Biol. Inorg. Chem., 2007, 12, 1015-1027. [ Links ]
9 Q.L. Zhang, J.H. Liu, J.Z. Liu, P.X. Zhang, X.Z. Ren, Y. Liu, Y. Huang and L.N. Ji, J. Inorg. Biochem, 2004, 98, 1405-12. [ Links ]
10 X.W. Liu, L.C. Xu and H. Li, H. Chao, K.C. Zheng and L.N. Ji, J. Mol. Struct, 2009, 920, 163-171. [ Links ]
11 D.S. Sigman, Acc. Chem. Res., 1986, 19, 180-186. [ Links ]
12 Y. Jenkins, A.E. Friedman, N.J. Turro and J.K. Barton, Biochemistry, 1992, 31, 10809-10816. [ Links ]
13 D.L. Carlson, D.H. Huchital, E.J. Mantilla, R.D. Sheardy and W.R. Murphy, J. Am. Chem. Soc., 1993, 115, 6424-6425. [ Links ]
14 A. Masood and D.J. Hodgson, Inorg. Chem, 1993, 32, 4839-4844. [ Links ]
15 B. Coyle, K. Kwanagh, M. McCann, M. Devereux and M. Geraghty, Biometals., 2003, 16, 321-329. [ Links ]
16 H. Qizhuang, Y. Jing, M. Hui and L. Hexing, Materials Letters, 2006, 60, 317-320. [ Links ]
17 J. Bolger, A. Gourdon, E. Ishow and J. P. Launay, Inorg. Chem., 1996, 35, 2937-2944. [ Links ]
18 H. Xu,K.C. Zheng,Y. Chen, Y.Z. Li, L.J. Lin,H. Li,P.X. ZhangandL.N. Ji, Dalton Trans., 2003, 11, 2260-2268. [ Links ]
19 C. Hiort, P. Lincoln and B. Norden, J. Am. Chem. Soc., 1993, 115, 3448-3454. [ Links ]
20 D. Greenwood, R. Snack and J. Peurtherer, Medical Microbiology: A Guide to Microbial Infections: Pathogenesis, Immunity, Laboratory Diagnosis and Control, 15th edn., Churchill Livingstone, Edinburgh, United Kingdom, 1997, p. 690. [ Links ]
21 V Reddy, N. Patil and S.D. Angadi, E. J. Chem., 2008, 5, 577-583. [ Links ]
22 SAS User's Guide: Statistics, Ver. 8, SAS 2001, SAS Institute, Cary, [ Links ]
23 H. Adams, R. Bastida, A. De Blas, M. Carnota, D.E. Fenton, A. Maci'as, A. Rodriguez and T. Rodriguez-Blas, Polyhedron, 1997, 16, 567-572. [ Links ]
24 W. Radecka-Paryzek and V. Patroniak, Polyhedron, 1994, 13, 21252128. [ Links ]
25 S. Liu, L.W. Yang, S.J. Rettig and C. Orvig, Inorg. Chem., 1993, 32, 2773-2778. [ Links ]
26 J.G. Liu, B.H. Ye, H. Li, Q.X. Zhen, L.N. Ji and Y.H. Fu, J. Inorg. Biochem., 1999, 76, 265-271. [ Links ]
27 D.H. Busch and J.C. Bailar Jr, J. Am. Chem. Soc., 1956, 78, 1137-1142. [ Links ]
28 M.M. Mashaly, H.F. El-Shafiy, S.B. El-Maraghy and H.A. Habib, Spectrochim. Acta A., 2005, 61, 1853-1869. [ Links ]
29 J. Bolger, A. Gourdon, E. Ishow and J.P. Launay, J. Chem. Soc., Chem. Commun., 1995, 17, 1799-1800. [ Links ]
30 J. Bolger, A. Gourdon, E. Ishow and J.P. Launay, Inorg. Chem., 1996, 35, 2937-2944. [ Links ]
31 J.E. Huheey, E.A. Keiter and R.L. Keiter, Inorganic Chemistry, Principle of Structure and Reactivity, Harper Collins College Publisher, New York, 1993, p. 1052. [ Links ]
32 A.B.P. Lever, Inorganic Electronic Spectroscopy, Elsevier, Amsterdam, 1986, p. 863. [ Links ]
33 F.A. Cotton and G. Wilkinson, Advanced Inorganic Chemistry, Wiley-Interscience Publication, New York, 1980, p. 1396. [ Links ]
34 W.J. Geary, Coord. Chem. Rev., 1971, 7, 81-122. [ Links ]
Received 9 May 2012
Revised 2 November 2012
Accepted 11 April 2013
* To whom correspondence should be addressed. E-mail: mesutgomleksiz@gmail.com














