Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Animal Science
versão On-line ISSN 2221-4062
versão impressa ISSN 0375-1589
S. Afr. j. anim. sci. vol.53 no.4 Pretoria 2023
http://dx.doi.org/10.4314/sajas.v53i4.05
Genetic diversity and population structure of Turkish Aseel chickens
F.T. Özbacer BulutI, #; Y. ÖzcensoyII; A. KocakayaIII; B. Yüceer ÖzkulIII; C. ÖzbeyazIII
IDepartment of Animal Breeding and Husbandry, Faculty of Veterinary Medicine, Tekirdag Namik Kemal University, Tekirdag, Türkiye
IIDepartment of Veterinary Genetics, Faculty of Veterinary Medicine, Sivas Cumhuriyet University, Sivas, Türkiye
IIIDepartment of Animal Husbandry, Faculty of Veterinary Medicine, Ankara University, Ankara, Türkiye
ABSTRACT
This study aimed to assess the genetic diversity, maternal origin, and population structure of Turkish Aseel chickens. The research was conducted on Aseel roosters and chicks older than one year. DNA was extracted from 96 Turkish Aseel feather samples collected from various regions of Türkiye for the study. Sample DNAs were amplified using specific primers for the D-loop region of the chicken mitochondrial DNA (mtDNA). The sequences were produced using the amplified DNA samples. The mtDNA D-loop regions were compared with the sequences of the same genetic area from other chicken breeds. Turkish Aseel chickens provided 34 haplotypes and 18 genuine polymorphisms across 41 distinct regions that were particular to the breed. According to the findings of the phylogenetic tree, Turkish Aseel chickens tend to establish clusters of haplogroups among themselves. Although most Turkish Aseel samples clustered individually into three clades, at least one sample has been discovered with five distinct clades. According to the phylogenetic tree and the Median Joining Network, it can be concluded that while many of the haplogroups of Turkish Aseel cluster among themselves, they have diverse maternal origins and lack breed-specific maternal lineages. This research is the first extensive study to examine genetic variability in Turkish Aseel chickens. Considering the lack of similar data on other Aseel varieties raised in different counties, the current work is a pioneering study with data on the genetic characterization of the Aseel breed.
Keywords: D-loop, genetic characterization, haplotype, mtDNA, Türkiye
Introduction
Various breeds of chickens are raised in Türkiye to produce meat, eggs, and other related products. Chicken types are also cultivated to participate in poultry-related hobbies (as ornamental or game birds). Despite the intensive import of commercial chicken breeds, some domestic animal gene sources have been secured for many years in Türkiye (Karaman & Kirdag, 2012). The Aseel is a chicken breed that is produced in many locations worldwide and is well-known for its diversity (Sarker et al., 2012; Mahmood et al., 2017; Rajkumar et al., 2017). A significant number of Aseel varieties are referred to by the name of the area in which they were produced (Rajkumar et al., 2017). It is conjectured that Aseel chickens were introduced as gamecocks to the Anatolian geography from South Asian nations such as India and Pakistan many centuries ago (Aldirmaz, 2020). By registering with federations or units linked with federations, they were grown intensively in various geographical regions of Türkiye (Atasoy et al., 2016).
The body structure of Turkish Aseel chickens is characterized by their compact and erect musculoskeletal system. The chicken has a straightened back and chest structures are short and broad. They have a strong beak that is curved towards the end, a thick neck, and an arched eye structure (Atasoy et al., 2016). The literature review and interviews with breeders reveal various morphological,
morphometric, and behavioural differences between the Aseel breeds raised in the regions and the Turkish Aseel chickens. The stubbornness of their structure and the fact that they cannot abandon fighting games are defining traits of this group. The studies conducted in Türkiye assessed physical features of Turkish Aseel chickens (Atasoy et al., 2016). There has been no genetic characterization of these birds, nor has any determination of their genetic or ancestral link with Aseel and other breeds produced in different geographic regions around the world been established.
Throughout history, there have been both natural and human-made selections in domestic chickens. Therefore, the diversified hybridizations lead to a rich gene pool in poultry (Quan et al., 2020). The population studies on genetic diversity in chickens determine the blood relations of breeds with each other and the geographical region where they were domesticated. Mitochondrial DNA (mtDNA) is one of the most studied target regions for determining genetic diversity in chickens (Teinlek et al., 2018; Kangayan et al., 2022; Phromnoi et al., 2022). The genome of mtDNA is maternally inherited. The observed mutation rate is faster in mtDNA than in nuclear DNA (nDNA) because mutations occurring in mtDNA are approximately 10-20 times higher than in nDNA, and mtDNA does not have a repair mechanism. The resulting mutation rate leads to very different variations in the base sequence of mtDNA (Özsensoy & Kurar, 2012). Specific target locations on mtDNA, such as the hypervariable D-loop, are commonly employed to study genetic diversity within and across breeds, to determine the phylogenetic link between species, and to ascertain the degree of relatedness between different breeds (Nishibori et al., 2003; Karaman & Kirdag, 2012; Özbaser et al., 2020; Godinez et al., 2021; Özarslan et al., 2021).
There are studies that have determined the origins of the breed of fighting roosters using mtDNA (Xiaoxu et al., 2015; Teinlek et al., 2018; Hata et al., 2021). Some of these studies were conducted on fighting breeds bred in China and Japan (Liu et al., 2006; Qu et al., 2009; Liu & Zhao, 2010; Xiaoxu et al., 2015). Qu et al. (2009) reported that Zhangzhou and Xishuangbanna fighting roosters in China originated from two haplogroups. Liu & Zhao (2010) reported that Chinese fighting roosters consisted of seven haplogroups. Although the Aseel is a gamecock that is widely bred globally, comprehensive studies on this breed's origin and genetic diversity have yet to be done.
This study was conducted to determine the genetic variations and haplotypes among other breeds and within the Turkish Aseel breed using mtDNA, an important genetic marker. In conjunction with the genetic data gathered, the mtDNA sequence analysis of the Turkish Aseel breed aimed to uncover the population's genetic structure and investigate the phylogenetic link with other chicken breeds.
Material and Methods
Ethical clearance for this research was granted by the Animal Care and Use Committee of Ankara University (Ethical clearance number 2017-25-206).
In this study, feather samples were collected from 50 female and 46 male birds of different ages that were grown in various enterprises registered with the Aseel Federation or organizations in nine cities in Türkiye (istanbul, Düzce, Sakarya, Sinop, Giresun, Samsun-Bafra, Trabzon, Antalya, and izmir), where Aseel rooster and chicken breeding is conducted. On a provincial scale, the samples were obtained from at least five distinct breeders in each city and no more than three samples from each breeder.
The processing of DNA samples was carried out in the laboratory of Sivas Cumhuriyet University, Faculty of Veterinary Medicine, Department of Veterinary Genetics. Genomic DNA from the collected feather samples was extracted using an automated nucleic acid isolation system (QuickGene-810) and a commercial kit (QuickGene, Fujifilm Life Science) according to manufacturers' protocol. The samples were stored at -20 °C until they were examined. The primers used (forward primer, F: 5' -> GGC TTG AAA AGC CAT TGT TG ->3'; reverse primer, R: 5'-> CCC CAA AAA GAG AAG GAA CC ->3') were made as reported by Muchadeyi et al. (2008).
Each polymerase chain reaction (PCR) was carried out in 30-uL reaction volume containing 1 * PCR buffer combination [0,5* potassium chloride (KCl) buffer and 0,5* ammonium buffer] (Ampliqon), 200 mM dNTP mix (Ampliqon), 1 U Ampliqon Red Taq polymerase (Ampliqon), 10 pmol of each primer, and ~10 ug of genomic DNA. PCR products were amplified in two steps using the Touchdown PCR profile (Don et al., 1991). The first step was initial denaturation at 95 °C for 15 min; followed by 12 cycles of denaturation at 94 °C for 1 min, annealing beginning at 66 °C with 0.5°C lowering for each cycle and ending at 60 °C for 1 min; and extension at 72 °C for 1 min. At the second step, 25 cycles of 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 1 min were applied. A final extension of 72 °C for 10 min was applied in all reactions.
PCR products were cleaned using the ExoSAP PCR purification kit (Thermofisher Scientific, USA) according to the manufacturer's instructions. The products were submitted to the sequencing
PCR process using the sequence kit (BigDye 3.1) and R primers after the purification procedure. After the sequence PCR was completed, the purification of the sequence PCR products was carried out using the gel filtering method using Sephadex. Then, PCR products were placed in the ABI Prism® 310 Genetic Analyzer and sequencing was performed. Comparative analyses were performed between obtained sequencing results and the publicly available NCBI GenBank data (Table 1).
The raw data of the sequences were loaded into UGENE 35.1 software (Okonechnikov et al., 2012), and the samples with poor sequence quality were removed. The mtDNA sequences were aligned and edited using the Clustal W Multiple Alignment method (Thompson et al., 1994) in the BIOEDIT 7.2.5 program (Hall, 1999), and polymorphisms were determined. The number of polymorphic regions, haplotype number, nucleotide diversity (Pi), haplotype diversity (Hd), average of nucleotide differentiation (k), testing of neutral theory (Tajima's D value, Fu's Fs statistical value, Fu and Li's D test, Fu and Li's F test), and mismatch distribution analyses were estimated using the DnaSP Ver.6.12.03 software suite (Rozas et al., 2017). For the identification of the Neighbour-Joining (NJ) tree and estimates of evolutionary divergence between sequences, they were constructed by using MEGA X software (Kumar et al., 2018) with the Kimura-2 parameter model (Kimura, 1980) and bootstrap test (1000 replicates) (Felsenstein, 1985). The Median Joining Network was generated using Network 10.1.0.0 (Bandelt et al., 1999) software only in Aseel samples and with other reference data in Table 1.
Results and Discussion
An automated DNA isolation method was used to extract DNA from 96 of the chicken feathers, and the extracted DNAs were amplified. PCR was conducted for the sequencing of the replicated mtDNA D-loop region samples, and the samples with inadequate sequence quality were removed. The remaining sequences of a total of 79 samples were verified and sorted. The polymorphisms of three Gallus gallus, one WL-2, Indian Aseel, and six indigenous chicken breeds (Table 1) with Turkish Aseel samples were identified and aligned. Here, the Gallus gallus gallus (GGG) sample was used as a reference; polymorphisms in Turkish Aseel samples are summarized in Table 2.
Polymorphisms were found in 41 different regions, 18 of which (242, 330, 355, 363, 367, 419, 422, 425, 426, 428, 432, 433, 436, 461, 462, 489, 500, and 503) were not present in other samples but only in Aseel samples (Table 2). Only one sample included five identified polymorphisms (167, 210, 212, 225, and 315). Five polymorphisms (167, 210, 212, 225, and 315) were observed in only one sample each. The polymorphism observed in the highest number of samples (n: 78) was the 296th nucleotide. Several polymorphisms in Turkish Aseels (nine regions) were observed in Indian Aseels as well.
A total of 34 haplotypes were identified in the studied Aseel population. While a total of 24 polymorphic regions were determined, two of these regions were singletons (296 and 315). Tajima's D value (1.71883), Fu and Li's D statistic (1.07455), and Fu and Li's F statistic (1.56900) were not statistically significant (P >0.10). In addition, Fu's Fs statistical value was calculated as -10.430 (Table 3).
To gain insight into the history of the population, mismatch distribution studies were conducted using the "Constant" (Figure 1A) and "Growth-Decline" (Figure 1B) models. The graph's green curves represent the anticipated, while the red curves represent the observed incompatibility.
According to estimates of evolutionary divergence between sequence values of Turkish Aseel samples, the genetic distances of the studied Turkish Aseel population were measured. Antalya 8 and Bafra16 samples were found to have the most significant genetic distance (0.051980) among the examined Turkish Aseel population samples. The next-farthest distance (0.051095) was the gap between the Bafra8 and Istanbul10 samples. It was found that many of the samples were genetically related (Figures 2 and 3).
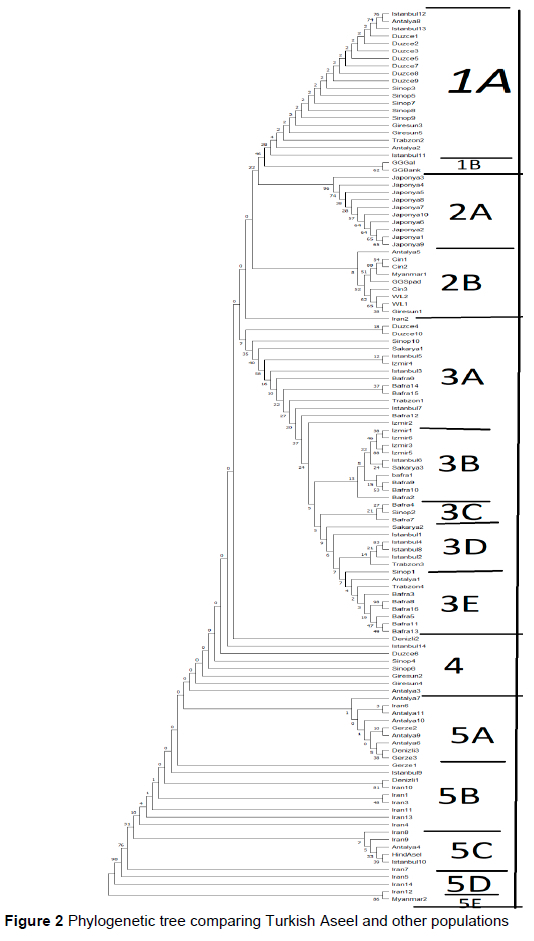
The phylogenetic tree of the samples from the Turkish Aseel population and other samples in Table 1 were examined using NJ tree constructions (Figures 2 and 3). Although most of the Turkish Aseel population was found among the three clade groupings, at least one was discovered in two distinct groups with individuals of other breeds. Therefore, it was determined that the Turkish Aseel was engaged in five distinct lineages. Even though the samples gathered from several provinces formed a group with the samples from their provinces or geographical areas, it was observed that a few geographically distant samples were collected together.
The Median Joining Network in which the frequencies of each haplotype in the Turkish Aseel mtDNA D-Loop region sequences were compared to the data of other chicken breeds is shown in Figure 4. Examining Figure 4 reveals that Turkish Aseels always formed a haplogroup cluster within themselves; nevertheless, a few samples (istanbul, Düzce, Bafra, Sinop, Giresun, Trabzon, and Antalya) were included in the examples of wild types, Japan, White Leghorn, China, and particularly Indian Aseels. Studies that targeted mtDNA have been conducted on various chicken breeds in several countries (Liu et al., 2006; Oka et al., 2007; Muchadeyi et al., 2008). For instance, 34 haplotypes were discovered in a study of 14 distinct Zimbabwean chicken groups using mtDNA D-loop sequencing (Muchadeyi et al., 2008). Liu et al. (2006) compared the origins and phylogeographic histories of 66 wild red forest hens (Galíus galíus) from Southeast Asia and China with 834 domestic hens (Galíus galíus domesticus) from throughout Eurasia by analysing mitochondrial DNA hypervariable segment I. According to this examination, there was no group, although several haplotypes were identified. Consistent with the findings of Muchadeyi et al. (2008) and Liu et al. (2006), several haplogroups were identified in Turkish Aseel hens in the present research. In addition to other chicken breeds, studies conducted on different cockfighting and gamecock breeds in China and Japan have identified many different haplotypes and haplogroups (Qu et al., 2009; Liu & Zhao, 2010; Xiaoxu et al., 2015). The large number of haplotypes and diversity of haplogroups in cockfighting and gamecocks, both in our study and other reported studies, is explained by the hypothesis that these breeds originated from multiple origins or evolved from wild and domestic chicken varieties in different regions (Qu et al., 2009).
Liu et al. (2006) suggested that the groups may have originated in various areas, including Greece, south and southwest China, possible neighbouring regions (Vietnam, Burma, and Thailand), and India. According to Meydan et al. (2016), Turkish and Iranian chicken breeds are closely related and may have originated from the same place (the Indian peninsula). In addition to the morphological similarities of the Turkish Aseel roosters and the Indian Aseel roosters, it is stated in historical sources that these roosters and chickens were brought from India and Pakistan many years ago and that they were cultivated in a controlled manner in the geography of Türkiye, particularly as competition roosters (Aldirmaz, 2020). Per this knowledge, genomic sequences of Aseel breeds growing in various geographic locations were addressed in this investigation. Although ten sequences have been uploaded to the gene bank of the Aseel breed from different geographies, nine are too short for phylogenetic comparisons; thus, no phylogenetic comparison can be made. One has the whole mtDNA sequence (Gene-Bank accession number: KP211418); hence genotypic comparisons of mt-DNA were only possible in one Aseel breed in the current study. In the polymorphism study conducted with a single Indian Aseel genotype, unlike other breeds, the Turkish Aseel polymorphisms were also seen in Indian Aseels (Table 2). Except for two (Antalya4, Istanbul 10), the phylogenetic comparison revealed that the Turkish Aseels belonged to a separate evolutionary group from the Indian Aseels (Figure 2).
In addition to the Gallus gallus gallus (GGG) sample used as a reference in the genetic investigation, polymorphisms (Table 2) were performed using Denizli, Gerze, WL, and other JFR samples from Türkiye's native chicken breeds. Polymorphisms existed in 41 distinct locations, 18 of which (242, 330, 355, 363, 367, 419, 422, 425, 426, 428, 432, 433, 436, 461, 462, 489, 500, and 503) were found solely in Aseel samples, which served as the study's subject matter (Table 2). Polymorphisms at nucleotide positions 296 and 342, which are polymorphisms also seen in other samples, were observed in all Turkish Aseel samples except samples 1 and 2, respectively. Polymorphisms in three regions (212, 246, and 315) only in GGS were observed in Aseel samples (1, 2, and 1 sample, respectively), while no other breeds were observed. In four locations (243, 256, 261, and 310), polymorphisms seen in both GGS and WL but not in other Turkish native breeds were detected in Aseel samples. No polymorphism was observed in Aseel samples in seven regions (67, 78, 199, 399, 457, 459, and 460) with polymorphisms according to the reference. Six sites (217, 281, 296, 306, 342, and 446) exhibiting polymorphism in Denizli and Gerze, two Turkish native breeds, were also detected in Aseel samples. Regionally and intra-regionally, nucleotide polymorphisms were found to be prevalent in chickens bred in various countries (Liu et al., 2006; Muchadeyi et al., 2008).
Muchadeyi et al. (2008) found a broad range of possible haplotype variations in the chicken breeds they investigated, ranging from 0.27 ± 0.12 to 0.78 ± 0.06. Generally, haplotype diversity and the number of polymorphic sites are reported to be low in local breeds (Meydan et al., 2016; Phromnoi et al., 2022). For example, in the study of Meydan et al. (2016) on native breeds bred in Turkey (Denizli and Gerze) and Iran (White Marandi, Black Marandi, Naked Neck, Common Breed, Lari, and West Azarbaijan), haploid diversities were found to be 0.24 ± 0.01 and 0.36 ± 0.02, respectively. It was also determined that there were 19 haplotypes and 24 polymorphic sites. In another study, the haplotype and nucleotide diversity of Khiew-Phalee Chickens (Gallus gallus), a Thai indigenous breed, were 0.597 and 0.00068, respectively (Phromnoi et al., 2022). Similar to the current study, it was stated that there were 24 haplotypes and six polymorphic sites in Nicobari chickens, an endemic breed raised from the Andaman and Nocobar Islands in India, in which haplotype diversity was 0.895 (Kangayan et al., 2022). Similarly, in the study conducted among Thai domestic chicken breeds (Pra-dhu-hang-dam, Leung-hang-khao, Chee, and Dang), it was reported that the average haplotype diversity was 0.8607 (Teinlek et al., 2018). In the current study population of Turkish Aseel chickens, 34 haplotypes were found in 24 different polymorphic sites. A high value of haplotype diversity (0.8971 ± 0.024) was also obtained in the Turkish Aseel. It is thought that the high haplotype diversity observed in this study was due to the multiple ancestral origins of the Aseel population.
In the current study, Tajima's D test showed an insignificant deviation from neutrality; its value was 1.71883 (Table 3). Similar to the result of the current study, in the domestic chicken breeds (Pra-dhu-hang-dam, Leung-hang-khao, Chee, and Dang) reared in Thailand, the total Tajima D test results were determined as insignificant deviation and positive values (except for the Dengue breed; - 0.00305) (Teinlek et al., 2018). In another study conducted on local Thai chicken breeds (Lueng-hang-khawo, Chee, Pradu-hang-dam, Kheaw-Paree, Betong, Decoy, Nin Kaset Nin, Dong-Tao Fighting chicken), although the results of the Tajima D test were insignificant similar to our study, the values were negative (-0.67) (Hata et al., 2021). The positivity of Tajima's D test value indicates that heterozygosity in the population provides a selective advantage, and a rapid increase in the population does not occur (Stephens et al., 2001). Statistically insignificance of Tajima's D test in studies indicates that the population is in neutral equilibrium, there is no natural selection in the neutral model, and the number of rare variations is low.
These numbers imply that the Aseel population has been stable for an extended period. Fu's Fs statistical value was found to be -10.430, which shows the existence of newly developing haplotypes and may be regarded as data that can explain the observation of 18 nucleotide polymorphisms (Table 2) in the Turkish Aseel population exclusively.
Examining the Median Joining Network of indigenous Aseel and other chicken populations reveals that most Turkish Aseel samples belong to a distinct group. However, specific haplotypes were in the middle of other chicken breed populations (Figure 4). The haplotypes were found at a high frequency in Turkish Aseel samples. Furthermore, a group of Aseel samples with only other haplotypes was detected on the right side of the centre. In addition, other Turkish native breeds formed a group with Turkish Aseel. Similar to the evolutionary tree, Japan and Iran were differentiated from other haplotypes by their groupings.
Geographically, Türkiye occupies a position of transition between Asia and Europe. Due to its location, Türkiye has historically served as a trade and migration route, resulting in the co-breeding of several animal breeds in Türkiye, as well as extensive hybridization and high genetic material flow across breeds due to human activities. That there is more than one haplogroup or different maternal origins in the Turkish Aseel chickens in the current study may be because of the location of Türkiye. Similar results have been reported in other animals, such as the dogs, in Türkiye (Özbacer et al., 2020).
Conclusions
The present study is the first to comprehensively describe the genomic diversity and population structure of Turkish Aseel chickens. Furthermore, this is one of the few studies on the Aseel breed, reportedly raised in many countries. When the genetic data are evaluated together, it can be inferred that the Turkish Aseel chicken samples were in a completely separate haplogroup from the races in other countries, except for a few, and that Turkish Aseel chickens could be a separate genotype. However, more Aseel varieties from distinct geographic regions and more detailed genotyping analyses are required to draw stronger conclusions.
Acknowledgements
We thank the owners of Aseel Roosters for their support on this study. This research was financially supported by the Ankara University Scientific Research Projects Coordinator (project no. 18B0239004). This study is dedicated to Fatih Atasoy, who was one of the planners of the study and passed away.
Authors' Contributions
All of the authors contributed to the conceptualization of this study; FTBÖ and AK collected the samples; YÖ conducted the laboratory analyses; YÖ, BYÖ, and CÖ performed the statistical analysis; FTÖB, YÖ, BYÖ, and CÖ interpreted the results and drafted the manuscript; FTÖB and YÖ reviewed and edited the article; and CÖ administered the project.
Conflicts of Interest Declaration
The authors of this manuscript have declared that there is no conflict of interest pertaining to this work.
References
Aldirmaz, M., 2020. Fight or competition? Folluk Dergisi, 34-46. [ Links ]
Atasoy, A., Yüceer, Ö.B. & Özbacer, F.T., 2016. Determination of some morphological features of Aseel cock and hens rearing in Turkey. Lalahan Hayvancilik Araçtirma Enst. Derg. 56, 56-62. https://dergipark.org.tr/tr/download/article-file/544582 [ Links ]
Bandelt, H.J., Forster, P. & Röhl, A., 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37-48. https://doi.org/10.1093/oxfordjournals.molbev.a026036 [ Links ]
Desjardins, P. & Morais, R., 1990. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J. Mol. Biol. 212, 599-634. https://doi.org/10.1016/0022-2836(90)90225-B [ Links ]
Don, R.H., Cox, P.T., Wainwright, B.J., Baker, K. & Mattick, J.S., 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res, 19, 4008. https://doi.org/10.1093/nar/19.14.4008 [ Links ]
Felsenstein, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evol. 39, 783-791. https://doi.org/10.2307/2408678 [ Links ]
Godinez, C.J.P., Dadios, P.J.D., Espina, D.M., Matsunaga, M. & Nishibori, M., 2021. Population genetic structure and contribution of Philippine chickens to the Pacific chicken diversity inferred from mitochondrial DNA. Front Genet. 12, 698401. https://doi.org/10.3389/fgene.2021.698401 [ Links ]
Hall, T.A., 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95-98. [ Links ]
Hata, A., Nunome, M., Suwanasopee, T., Duengkae, P., Chaiwatana, S., Chamchumroon, W., Suzuki, T., Koonawootrittriron, S., Matsuda, Y., & Kornsorn Srikulnath, K., 2021. Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens. Sci. Rep. 11, 2035. https://doi.org/10.1038/s41598-021-81589-7 [ Links ]
Kangayan, M., Kumar De, A., Bhattacharya, D., Tamilvanan, S., Ponraj. P., Alyethodi, R.R., Sunder, J., Bala, P.A, Kundu A. & Chakurkar E.B., 2022. Genetic structure, population diversity and ancestry of Nicobari fowl based on mtDNA complete D-loop sequences. J. Genet. 101:31. https://doi.org/10.1007/s12041-022-01372-z [ Links ]
Karaman, M. & Kirdag, N., 2012. Mitochondrial DNA D-loop and 12S regions analysis of the long-crowing local breed Denizli fowl from Turkey. Kafkas Univ. Vet. Fak. Derg. 18, 191-196. https://doi.org/10.9775/kvfd.2011.5248 [ Links ]
Kimura, M., 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111-120. https://doi.org/10.1007/BF01731581 [ Links ]
Kirdag, N., 2007. Application of molecular techniques to poultry phylogenetic studies. Masters Thesis, Kahramanmaras Sütçü imam Üniversitesi Fen Bilimleri Enstitüsü. Türkiye. [ Links ]
Komiyama, T., Ikeo, K. & Gojobori, T., 2003. Where is the origin of the Japanese gamecocks? Gene. 317, 195-202. https://doi.org/10.1016/S0378-1119(03)00703-0 [ Links ]
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K., 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547-1549. https://doi.org/10.1093/molbev/msy096 [ Links ]
Liu, Y.P., Wu, G.S., Yao, Y.G., Miao, Y.W., Luikart, G., Baig, M., BejaPereira, A., Ding, Z.L., Palanichamy, M.G. & Zhang Y.P., 2006. Multiple maternal origins of chickens: Out of the Asian jungles. Mol. Phylogenet. Evol. 38, 112-9. https://doi.org/10.1016/j.ympev.2005.09.014 [ Links ]
Liu, W.Y. & Zhao, Chun-Jiang., 2010. Comprehensive genetics analysis with mitochondrial DNA data reveals the population evolution relationship between Chinese gamecocks and their neighboring native chicken breeds. Asian J. Anim. Vet. Adv. 5, 388-401. https://doi.org/10.3923/ajava.2010.388.401 [ Links ]
Mahmood, S., Rehman, A., Khan, M., Lawal, R. A. & Hanotte, O., 2017. Phenotypic diversity among indigenous cockfighting (Aseel) chickens from Pakistan. J. Anim. Plant Sci. 27, 1126-1132. http://www.thejaps.org.pk/docs/v-27-04/10.pdf [ Links ]
Meydan, H., Jang, C.P., Yildiz, M.A. & Weigend, S., 2016. Maternal origin of Turkish and Iranian native chickens inferred from mitochondrial DNA D-loop sequences. Asian-Australas. J. Anim. Sci. 29, 1547-1554. https://doi.org/10.5713/ajas.15.1060 [ Links ]
Muchadeyi, F.C., Eding, H., Simianer, H., Wollny, C.B.A., Groeneveld, E. & Weigend, S., 2008. Mitochondrial DNA D-loop sequences suggest a southeast Asian and Indian origin of Zimbabwean village chickens. Anim. Genet. 39, 615-622. https://doi.org/10.1111/j.1365-2052.2008.01785.x. [ Links ]
Nishibori, M., Hanazono, M., Yamamoto, Y., Tsudzuki, M. & Yasue, H., 2003. Complete nucleotide sequence of mitochondrial DNA in White Leghorn and White Plymouth Rock chickens. Anim. Sci. J. 74, 437-439. https://doi.org/10.1046/j.1344-3941.2003.00136.x [ Links ]
Nishibori, M., Shimogiri, T., Hayashi, T. & Yasue, H., 2005. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim. Genet. 36, 367-375. https://doi.org/10.1111/j.1365-2052.2005.01318.x. [ Links ]
Oka, T., Ino, Y. & Nomura, K., 2007. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim. Genet. 38, 287-93. https://doi.org/10.1111/j.1365-2052.2007.01604.x [ Links ]
Okonechnikov, K., Golosova, O., Fursov, M. & UGENE team, 2012. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 28, 1166-1167. [ Links ]
zarslan, B., Atasoy, F., Erdogan, M., Yüceer Özkul, B. & Özbacer, F.T., 2021. Evaluation of some morphological and genetic characteristics of Çatalburun dog breed. Turkish J. Vet. Anim. Sci. 45, 649-656. https://doi.org/10.3906/vet-2012-20 [ Links ]
Özbacer, F.T., Atasoy, F., Erdogan, M., Yüceer, Özkul. B. & Özarslan, B., 2020. Morphological and genetic characteristics of Zerdava, a native Turkish dog breed. Kafkas Univ. Vet. Fak. Derg. 26, 617-623. https://doi.org/10.9775/kvfd.2020.24004 [ Links ]
Özcensoy, Y. & Kurar, E., 2012. Marker systems and applications in genetic characterization studies. J. Cell Mol. Biol. 10, 11-19. [ Links ]
Phromnoi, S. Lertwatcharasarakul, P. & Phattanakunanan S., 2022. Genetic diversity and phylogenetic analysis of Khiew-Phalee chickens (Thailand) based on mitochondrial DNA cytochrome b gene sequences. Biodivers. 23, 750-756. https://doi.org/10.13057/biodiv/d230220 [ Links ]
Rajkumar, U., Haunshi, S., Paswan, C., Raju, M.V.N., Rama Rao, S.V. & Chatterjee, R.N., 2017. Characterization of indigenous Aseel chicken breed for morphological, growth, production, and meat composition traits from India. Poult. Sci. 96, 2120-2126. https://doi.org/10.3382/ps/pew492. [ Links ]
Rozas, J., Ferrer-Mata A., Sánchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E. & Sánchez-Gracia, A., 2017. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 34, 3299-3302. https://doi.org/10.1093/molbev/msx248 [ Links ]
Sarker, M.J.A., Bhuiyan, M.S.A., Faruque, M.O., Ali, M.A. & Lee, J.H., 2012. Phenotypic characterization of Aseel chicken of Bangladesh. Korean J. Poult. Sci. 39, 9-15. https://doi.org/10.5536/KJPS.2012.39.L009 [ Links ]
Stephens, J.C., Schneider, J.A, Tanguay, D.A, Choi, J., Acharya, T., Stanley, S.E, Jiang, R., Messer, C.J, Chew, A, Han, J.H, Duan, J, Carr, J.L, Lee, M.S, Koshy, B., Kumar, A.M, Zhang, G., Newell, W.R, Windemuth, A., Xu, C., Kalbfleisch, T.S, Shaner, S.L., Arnold, K., Schulz, V., Drysdale, C.M., Nandabalan, K., Judson, R.S., Ruan~o, G. & Vovis G.F., 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science, 293, 489-493. https://doi.org/10.1126/science.1059431 [ Links ]
Taskesen, H.O., 2010. Mitochondrial DNA D-Loop polymorphism in Denizli chicken population. Masters Thesis, Ankara Üniversitesi Fen Bilimleri Enstitüsü, Türkiye. [ Links ]
Teinlek, P., Siripattarapravat, K. & Tirawattanawanich, C., 2018. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian-Australas J. Anim. Sci. 31, 804-811. https://doi.org/10.5713/ajas.17.0611 [ Links ]
Thompson, J.D., Higgins, D.G. & Gibson, T.J., 1994. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673-4680. https://doi.org/10.1093/nar/22.22.4673 [ Links ]
Quan, J., Cai, Y., Yang, T., Ge, Q., Jiao, T. & Zhao, S., 2020. Phylogeny and conservation priority assessment of Asian domestic chicken genetic resources. Glob. Ecol. Conserv. 22, e00944. https://doi.org/10.1016/j.gecco.2020.e00944 [ Links ]
Qu, L.J., Li, X.Y. & Yan, N., 2009. Genetic relationships among different breeds of Chinese gamecocks revealed by mtDNA variation. Asian-Aust. J. Anim. Sci. 22, 1085-1090. https://doi.org/10.5713/ajas.2009.80660 [ Links ]
Xiaoxu, J., Junxian, K., Xiujun, T., Mengjun, T., Yanfeng, F., Rong, G., Yushi, G. & Yijun, S.U., 2015. Mitochondrial DNA control region polymorphisms in Henan gamecocks and evolutionary relatedness with other four gamecock breeds. J. Anhui Agric. Univ. 42, 726-729. https://doi.org/10.13610/j.cnki.1672-352x.20150825.003 [ Links ]
Submitted 9 March 2023
Accepted 3 May 2023
Published 26 August 2023
# Corresponding author: ftozbaser@nku.edu.tr














