Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.53 n.3 Pretoria 2023
http://dx.doi.org/10.4314/sajas.v53i3.09
Association study of polymorphisms in miRNA-1687 with growth traits of chickens
J.Z. ShiI; Y.W. WangII; T.J. WangIII; J. JiI; G.R. SunIV, #; L.G. YaoI, #
INanyang Normal University, 473061, Nanyang, China
IIZhengzhou University, Zhengzhou, 450001, China
IIINanyang Vocational College of Agriculture, 473000, Nanyang, China
IVHenan Agricultural University, Zhengzhou, 450002, China
ABSTRACT
Polymorphisms within microRNAs can lead to phenotypic variations in organisms. The purpose of this research was to investigate the potential impact of the pre-miR-1687 single nucleotide polymorphism (SNP) on the economic characteristics of weight and body size in chickens. The SNP was genotyped using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. A linear mixed model was utilized to analyse the associations between the SNP and chicken body size and growth traits. The SNP in the pre-miR-1687 gene was correlated with F2 chicken body weight (BW) at birth and at 4, 6, 8, and 10 weeks of age. The SNP in the gga-miR-1687 gene was correlated with shank length, shank girth, pectoral angle (at 4 weeks), and pelvic breadth (at 8 weeks). Different BW genotypes were observed in the studied flocks. The changes in the secondary structure of pre-miR-1687 and in the free energy values were estimated using online M-fold software. The results serve as a helpful resource for subsequent research on the mechanisms and functions of miRNAs. In addition, the study provides a credible basis for the application of biomolecular technology in poultry breeding.
Keywords: correlation analysis; F2 chicken population; growth trait; pre-miR-1687; single nucleotide polymorphism
Introduction
MicroRNAs (miRNAs), which are widely expressed in many organisms and tissues in animals and humans, are a novel group of highly conserved, small noncoding RNAs (ncRNAs) that are approximately 18 to 22 nucleotides (nt) in length and act as master regulators of gene expression at the posttranscriptional stage via RNA interference pathways (Kabekkodu et al., 2018; Rani and Sengar, 2022; Salim et al., 2022). They are major posttranscriptional regulators and perform various functions within cells, including modulation of gene expression. MiRNAs can regulate and control one-third of all protein-coding genes by base-pairing to target mRNAs complementary to the 3' untranslated region (3' UTR), resulting in mRNA degradation and inhibition of mRNA translation (Sand et al., 2009; Salim et al., 2022). MicroRNAs are transcription products derived from endogenous DNA transcripts, and the biogenesis of miRNAs begins at internuclear primary transcripts that may or may not code proteins and are present in either intergenic regions or overlapping genes. In the cell nucleus, almost all primary transcripts are biologically processed into hairpin-like structures, partially duplexed pre-miRNAs (precursor miRNAs), by the RNase III Drosha; pre-miRNAs are usually 55 nt to 80 nt in length and are sent to the cytoplasm by Exportin-5 (a shuttle protein) with the aid of the G-protein, Ran, at the time of bioprocessing. The pre-miRNAs are identified and cleaved by Dicer RNaseIII. They are precisely processed into approximately 18- to 22-nt-long miRNA/miRNA* double strands and bind to the RNA-binding protein (RBP) of the regulatory factor transactivation response element (TRE) in the cytoplasm.
The RNA-induced silencing complex (RISC) promotes the conjugation of one chain of the miRNA double strand as a mature miRNA to match the matching mRNA sequence in the 3' UTR of a target gene; the other chain of the double strand is degraded (Wang et al., 2013; Li et al., 2014).
Accumulating evidence confirms that miRNAs are associated with an extremely wide range of biological processes, for example, sperm motility (Kumar et al., 2015), embryonic development (Collignon, 2007; Yuan et al., 2016), haemopoiesis (Lazare et al., 2014; Kotaki et al., 2017; Rasko & Wong, 2017), fat deposition (Zhang et al., 2016; Qiang et al., 2018), cell differentiation (Martin et al., 2016; Zhao et al., 2020), cell proliferation (Wang et al., 2017; Du et al., 2020), oocyte maturation (Song et al., 2016; Zhang et al., 2019a), ovarian follicular development (Imbar and Eisenberg, 2014; Maalouf et al., 2016; Zhang et al., 2019b), metabolism (Vienberg et al., 2017; Alamoudi et al., 2018), apoptosis (Fan et al., 2016; Akkafa et al., 2018), stem cell maintenance (Mehta et al., 2015; Luinenburg & de Haan, 2020), and skeletal muscle growth (Diniz and Wang, 2016; Wang et al., 2018).
Single-nucleotide polymorphisms (SNPs) located on miRNA-binding sites (miR-SNPs) have an effect on gene expression. Normally, SNPs influence miRNA function by altering miRNA expression levels and relevant targets. Common SNPs occurring in miRNAs can regulate the transcription of pri-miRNA transcripts and the stability or biological processing of pre-miRNAs or pri-miRNAs. The sites of miRNA targets and processing machinery might potentially affect the expression of many genes and pathways and might profoundly interfere with the function of miRNAs, resulting in either an increase or a decrease in mature miRNA levels, including individual phenotypic variations and disease susceptibility (Duan et al., 2007; Jazdzewski, 2008; Mencia et al., 2009).
Chickens are among the most economically, culturally, and socially important poultry species. Previous research has demonstrated the significant effectiveness of a SNP (AB604331, g.420 C>A) occurring in the gene of the cholecystokinin type A receptor for improving the growth characteristics of Amakusa x Daioh cross chickens (Momoi et al., 2021). The mutation, c.-652 C>T, in the promoter region of the ubiquitin carboxyl-terminal hydrolase-L1 gene was identified as being relevant to goose growth performance (Wang et al., 2021). A research report found that the SNPs, G244A and A239G, in exon 2 of the growth hormone secretagogue receptor were strongly correlated with feed intake, growth characteristics, and expression of the growth hormone secretagogue receptor gene, growth hormone, and the endogenous growth hormone receptor ghrelin (GHRL) mRNAs (El-Magd et al., 2016). SNPs (g.69307744 C>T, g.69340192 G>A, and g.69355665 T>C) had a marked impact on carcass characteristics, such as carcass and semi-eviscerated weight. This evidence showed that the SNP in the TBC1D1 gene was strongly associated with carcass traits in poultry flocks (Wang, 2014). The growth traits of chickens are among their most important economically important traits.
In the present study, we screened and identified a novel SNP located in the precursor of Gallus gallus miR-1687 (gga-miR-1687) and researched the correlation between the SNP and growth traits, including body weight (BW) and body indices, at multiple growth and developmental stages of chickens of the Gushi x Anka F2 reference family. The influence of polymorphisms in the pre-miR-1687 gene on the secondary structure and energy of pre-miRNA was also predicted and analysed. In general, the data identified the potential application value of miR-1687 in regulating chicken growth.
Materials and Methods
The related poultry experiments were conducted according to the rules and requirements of the Animal Care & Use Committee of Henan Agricultural University, China (approval ID: 11-0085).
The study design was adapted from Han et al. (2011). Briefly, a Gushi x Anka F2 resource population was used that included a total of 860 chickens. The study population previously established by the researchers of the Henan Innovative Engineering Research Center of Poultry Germplasm Resource (Han et al., 2012; Shi and Sun, 2017) was used as the source of experimental subjects for research. Specifically, the resource population of F2 chickens was derived by crossing Gushi chickens and Anka broiler chickens. Gushi chickens, a typical, slow-growing, native Chinese chicken breed, exhibit excellent meat characteristics. In contrast, Anka broiler chickens are large-bodied, fast-growing broiler chickens. In other words, a slow-growing breed (Gushi chickens) was crossed with a fast-growing breed (Anka broiler chickens). The F0 chickens (42 grandparents) were derived from two groups: one group (two reciprocal cross families) was generated by mating two Gushi roosters with 12 Anka broiler hens, and the other group (four cross families) was generated by mating four Anka broiler roosters with 24 Gushi hens. As noted earlier, this F2 resource population was established by intercrossing 63 F1 parental hens and seven F1 parental cockerels.
All the experimental chickens experienced identical breeding conditions and were raised in cages until 12 weeks of age, with free access to food and water. The 860 F2 chickens were all humanely slaughtered at 84 days of age. All animal experimental procedures used complied with the related animal care protocol.
The BW and body size traits of each chicken were measured accurately and recorded every two weeks from week 0 (hatching) to 12 weeks of age (i.e., weeks 0, 2, 4, 6, 8, 10, and 12). The primary chicken body size parameters measured at every different stage of growth and development (4, 8 and 12 weeks) were slanting body length, shank length, shank girth, chest depth, chest breadth, breastbone length, pectoral angle, and pelvic breadth.
A total of 860 genomic DNA samples of the Gushi x Anka F2 chicken reference family were extracted from venous blood by adding the anticoagulant ethylenediaminetetraacetic acid (EDTA) using the phenol-chloroform method with a commercial DNA extraction kit (Tiangen Biotechnology Co. Ltd., Beijing). Considering the standards for genotyping using mass array matrix-assisted laser desorption and ionization/time-of-flight mass spectrometry (MALDI-TOF MS), the exact concentrations of the prepared DNA samples were measured, and the samples were stored at -80 °C for subsequent research. One hundred genomic DNA samples from individual F2 chickens were chosen at will, and equivalent amounts of the samples of each genomic DNA with the same working concentration were pooled. The sequence of the chicken pre-miR-1687 gene (GenBank: MI0007421), which was located on chromosome 20, was acquired from the online miRBase database (version 19.0). The pooled DNA for the F2 resource population was sequenced by Shanghai Sangon Biotechnology Company to scan for gene polymorphisms. A pair of PCR amplification primers (F: 5'- GCTGATGGTGTCGCGGTGAGC -3'; R: 5'- CTGCATAAAGATGGGAGAAG -3') for sequencing was designed on the basis of the location in the genome (NC_006107.3) to detect SNPs in the precursor region of miR-1687. The primers were applied to amplify the gene sequence, including that of pre-miR-1687. The DNA samples were delivered to a reputable testing company for detecting SNPs by mass array MALDI-TOF MS (Sequenom Inc., USA).
In the Gushi x Anka F2 chicken population, the rs15179830 (+ 90 bp T>G) SNP within pre-miR-1687 was genotyped using a MassARRAY-iPLEX Gold system. SNP genotyping was performed using a pair of specific primers for amplified sequences and one single-base extension primer designed by Professional Assay Design software (version 3.1). The upstream primer sequence for the pre-miR-1687 SNP was 5'-ACGTTGGATGTGAACAGCAACACAGCTAGG-3'; the downstream PCR amplification primer sequence for the SNP was 5'-ACGTTGGATGAGCAACTTCTTTGCTGGCTG-3'; and the single-base extension primer was 5'-AGTGACTGCAGCATAAAAA-3'. The SNP was genotyped by MALDI-TOF MS as described in the operation manual. The genotyping data for the peak area and call rate were collected. The alleles were designated automatically by the software package offered by the product manufacturer.
In chickens, due to the rs15179830 SNP with T>G alleles in pre-miR-1687, the most stable secondary structure of pre-mir-1687 with the lowest free energy was computed by the online M-fold web server. The absolute value of the free energy difference between the two different alleles (T/G) of miRNA-1687 was calculated as a parameter to evaluate the effect on the secondary structure of gga-miR-1687.
A statistical data analysis software program (SPSS 20.0) was applied for correlation analysis of the relevant data for the SNP of pre-miR-1687 and the growth and development traits of the Gushi x Anka F2 resource population birds. The association between gene polymorphisms and the F2 chicken population-associated economic characteristics was analysed by using a biostatistical program with an established linear mixed model. The model was applied to analyse and evaluate the correlation between SNPs and growth traits. In this data analysis, the mixed linear model equation was:
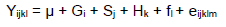
Yijkl in the equation represents the measured value, μ represents the overall average, Gi represents the fixed effects of the three genotypes (i = 3: TT, TG, GG), fl represents the random effect of the resource population (l = 7, 7 reference families), sj represents the fixed effect of sex (j = 2), Hk represents the hatch fixed effect (k = 2), and eyklm represents the random error. A P value <0.05 was recognized as statistically significant.
Results
The results for the polymorphism were confirmed using PCR amplification-based sequencing by comparison with the released chicken genome DNA sequence, which indicated the existence of a T>G mutation at +90 bp in pre-miR-1687 in the F2 resource population (Figure 1). The obtained mass spectrograms of three variant genotypes, namely, GG, GT, and TT, are shown in Figure 2. In this F2 resource population, the allele frequencies within the SNP in the pre-miR-1687 gene were 0.685 for G and 0.315 for T. Similarly, the genotype frequencies were 0.417, 0.537, and 0.046 for TT, GT, and GG, respectively.
The alleles of the pre-miR-1687 gene SNP can give rise to changes in the secondary structure of the miR-1687 RNA, as predicted by the online software program, M-fold. The prediction result showed that the SNP (+90 bp T>G) variation located in the precursor domain of pre-miR-1687 could generate one base pairing mismatch, leading to the production of a novel RNA ring-like protrusion in the secondary structure of the base mutant, along with a concurrent change in free energy (Figure 3).
To study the effect of the rs15179830 SNP in pre-miR-1687 on chicken phenotypic traits, an association analysis of the new SNP with the weight of chickens at different growth times (at birth and at 2 w, 4 w, 6 w, 8 w, 10 w, and 12 w of age) (Table 1). The pre-miR-1687 SNP gene had an impact (P <0.05) on BW at 0, 2, 4, 6, 8, and 10 weeks of age in the Gushi x Anka F2 chicken resource population. The F2 chickens with the TT genotype were the heaviest in terms of BW from 0-12 w. The results of the association analysis of different rs15179830 genotypes with chicken body size are presented in Table 2. The new mutation had an indirect influence on the economic characteristics (mainly including weight and body size) of the chickens at different growth and developmental stages.
Discussion
Single-stranded RNAs are short ncRNA molecules that regulate genes in a specific sequence. MicroRNAs perform vital functions in regulating gene expression by promoting the degradation of mRNA and inhibiting transcription. SNPs in miRNA genes can change the characteristics of miRNAs by changing the expression level of miRNAs or affecting the maturity of miRNAs. An increasing number of studies have focused on the functions of microRNAs in biological processes in recent years. Increasing evidence suggests that SNPs occurring in miRNA genes or their targets might affect the economic characteristics of livestock and poultry (Li et al., 2015). For example, more than twenty SNPs were screened for their association with days to 100 kg and birth weight in 600 Yorkshire pigs. One locus (location: 46 226 512 bp) located on SSC12 was assessed to affect both birth weight and age at 100 kg. According to their roles, the DOCK7 gene and NSRP1 gene are considered to be the most promising candidate genes associated with growth characteristics (Wu, 2021). In Hu sheep, one new SNP (g. 4819 A>G) in LIPE was found in intron 4 of the noncoding region of the fragmented gene, and this SNP was strongly related to the intramuscular fat content. The expression level of LIPE in the longissimus dorsi muscle was negatively correlated with intramuscular fat content. With increasing LIPE expression, the intramuscular fat content decreased.
These results showed that the SNP (g.4819 A>G) in the LIPE gene was promising as a molecular genetic marker for breeding Hu sheep with better intramuscular fat content (Kong et al., 2022). The missense SNP in HSD17B12 predicted to affect protein function was correlated with earlier submission for seasonal breeding, perhaps on account of earlier resumption of cyclicity postpartum in dairy cows (Juengel et al., 2022). In the cross of Nellore beef x Bos Taurus cattle, the SNP (g. 98 535 683 A>G) located in the BTAU7 calpastatin gene had an effect on beef quality traits. These results showed its application value for genetic breeding improvement of beef cattle for meat quality traits (Enriquez-Valencia et al., 2017). In Landrace and Yorkshire pigs, four notable SNPs (SSC6.149876737, 149876507, SSC8.54567459, SSC11.33043081) were discovered to be closely associated with growth performance. In addition, DOCK7, a functional candidate gene with potential application value, was recommended for selective breeding programs for BW and backfat thickness in Landrace and Yorkshire pigs (Yang et al., 2019). Analysis of the differences in expression at the transcriptional level for the SNP (g. 265 T>C) from FASN in semimembranous muscle tissue and backfat tissue of Italian Duroc (ID) pigs and Italian Large White (ILW) pigs was performed. The results showed that the transcription level of the fatty acid synthase (FASN) gene could be regarded as a molecular marker to identify the level of fat deposition in the ID pig breed (Braglia et al., 2014). One SNP (g.358 A>T) in the intronic location of the cluster-of-differentiation antigen-9 (CD9) molecule of Frieswal (HF x Sahiwal) crossbred bulls was strongly correlated with sperm content and sperm motility percentage (Kumar et al., 2015). One SNP (c. *7750 G>A) occurring in the miRNA-binding site of the epidermal growth factor receptor (EGFR) gene 3' UTR was related to egg production. The expression level of the EGFR gene may affect egg productivity in the Yangzhou goose breed (Alsiddig et al., 2022). The increase in BW and body size, which are important economic traits, is a result of the influence on production performance in chickens. The growth traits of chickens have a strong genetic background (Zerehdaran et al., 2004). Growth characteristics are important economic characteristics in poultry breeding.
The main purpose of the current study was to screen and evaluate the SNPs in pre-miR-1687 by making use of the Gushi x Anka F2 chicken resource population to determine if the SNPs were related to growth performance. The test method consisted of sequencing pre-miR-1687 by using a set of DNA samples from different F2 chicken individuals to identify the SNPs. We predicted the secondary structure and the free energy values of different alleles (T/G) of pre-miR-1687 by using online M-fold software. The SNPs altered the secondary structure and free energy of pre-miR-1687. The polymorphism, rs15179830, was identified in the pre-miR-1687 gene and was related to BW and pectoral angle in the chicken population. Correlation analysis was carried out for the SNP and phenotypic traits (growth traits) in F2 chickens. The Bonferroni-corrected model was used to decrease incorrect associations in the data analysis. The SNP was demonstrated to have a strong influence on the BW of chicks at 4, 6, 8, and 10 weeks and the pectoral angle at 4 w. In summary, the findings provide hard evidence that the gene polymorphism in pre-miR-1687 is associated with growth performance and is an available candidate gene molecular marker for breeding to effectively improve economic performance. We evaluated the effects of pre-miR-1687 in the Gushi x Anka F2 resource chicken population; correlation analyses of the SNP with growth characteristics have not been previously reported. In the present research, the results of the analysis confirmed that rs15179830 was strongly associated with birth weight, BW, at 4, 6, 8, and 10 weeks and PA at 4 w. The SNPs were strongly associated with diverse genotypes and growth traits.
Moreover, the genotypes GG, GT and TT were strongly related to BW in the order TT>GT>GG; T was a superior allele. The results showed that the T allele promoted the increase in BW, while the G allele was unfavourable for the increase in BW. The main aim of the current research was to identify the influence of the pre-miR-1687 SNP on chicken growth characteristics, and the findings enrich the knowledge and understanding of poultry genetic breeding technology, facilitating the development of improved genetic breeding schemes (Li et al., 2012). Therefore, the TT genotype of the SNP of the pre-miR-1687 gene was selected as the molecular genetic marker for improving BW in marker-assisted selection for poultry breeding.
Conclusion
Our data suggest that pre-miR-1687 might participate in regulating growth and development, implying that pre-miR-1687 may be a candidate gene correlated with chicken growth performance. Therefore, the SNP (rs15179830 T>G) of pre-miR-1687 is a potential, novel, molecular genetic marker that can be used for the selection of excellent growth and development traits in chickens. The results will be a helpful resource for further work on miRNA biological processes and functions and provide reference data for the application of molecular breeding technology in poultry genetics and breeding.
Acknowledgements
This study was supported by the Technology Innovation Team Project of Universities in Henan Province (20IRTSTHN024), the National Natural Science Foundation of China (Grant nos. 31870917, 31802185), the Henan Science and Technology Research Project (222102110260), the Key Scientific Research Projects of Higher Education Institutions in Henan Province (22A180026), and the NanYang Science and Technology Research Project (KJGG136).
Author Contributions
Conceptualization, G.R. Sun; methodology, G.R. Sun and J.Z. Shi; software, Y.W. Wang; validation, J.Z. Shi, T.J. Wang, and J. Ji; formal analysis, J.Z. Shi; investigation, G.R. Sun; resources, G.R. Sun; data curation, G.R. Sun; writing-original draft preparation, J.Z. Shi; writing-review and editing, J. Ji; visualization, J.Z. Shi; supervision, L.G. Yao; project administration, L.G. Yao; funding acquisition, L.G. Yao. All authors have read and agreed to the published version of the manuscript.
Conflict of interest declaration
All authors jointly declare that there is no conflict of interest in this study.
References
Akkafa, F., Koyuncu, i., Temiz, E., Dagli, H., Dïlmec, F. & Akbas, H., 2018. miRNA-mediated apoptosis activation through TMEM 48 inhibition in A549 cell line. Biochem. Biophys. Res. Commun. 503, 323-329. doi:10.1016/j.bbrc.2018.06.023 [ Links ]
Alamoudi, A.A., Alnoury, A. & Gad, H., 2018. miRNA in tumour metabolism and why could it be the preferred pathway for energy reprograming. Brief. Funct. Genomics. 17, 157-169. doi:10.1093/bfgp/elx023 [ Links ]
Alsiddig, M., Badri, T., Widaa, H., Li, B., Shigang, Y., Chen, J. & Liu, H., 2022. A SNP at MicroRNA binding site of epidermal growth factor receptor 3' untranslated region associated with Yangzhou geese egg production. Animal. Gene. 23. doi 10.1016/j.angen.2021.200123 [ Links ]
Bartel D.P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116(2), 281-297. doi:10.1016/s0092-8674(04)00045-5 [ Links ]
Braglia, S., Zappaterra, M., Zambonelli, P., Cornelia, M., Dall'Olio, S. & Davoli, R., 2014. Analysis of g.265T>C SNP of fatty acid synthase gene and expression study in skeletal muscle and backfat tissues of Italian Large White and Italian Duroc pigs. Livest. Sci. 162, 15-22. doi 10.1016/j.livsci.2014.01.014 [ Links ]
Collignon, J. 2007. miRNA in embryonic development: the taming of Nodal signaling. Dev. Cell. 13, 458-460. doi 10.1016/j.devcel.2007.09.012 [ Links ]
Diniz, G.P. & Wang, D.Z., 2016. Regulation of skeletal muscle by microRNAs. Compr. Physiol. 6, 1279-1294. doi 10.1002/cphy.c150041 [ Links ]
Du, B., Liu, X., Khan, A., Wan, S., Guo, X., Xue, J. & Fan, R., 2020. miRNA-183 approximately 96 approximately 182 regulates melanogenesis, cell proliferation, and migration in B16 cells. Acta. Histochem. 122, 151508. doi 10.1016/j.acthis.2020.151508 [ Links ]
Duan, R., Pak, C. & Jin, P., 2007. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet.16, 1124-1131. doi 10.1093/hmg/ddm062 [ Links ]
El-Magd, M.A., Saleh, A.A., Abdel-Hamid, T.M., Saleh, R.M. & Afifi, M.A., 2016. Is really endogenous ghrelin a hunger signal in chickens? Association of GHSR SNPs with increase appetite, growth traits, expression, and serum level of GHRL and GH. Gen. Comp. Endocrinol. 237, 131-139. doi 10.1016/j.ygcen.2016.08.016 [ Links ]
Enriquez-Valencia, C.E., Pereira, G.L., Malheiros, J.M., de Vasconcelos Silva, J., Albuquerque, L.G., de Oliveira, H.N., Chardulo, L.A.L. & Curi, R.A., 2017. Effect of the g.98535683A>G SNP in the CAST gene on meat traits of Nellore beef cattle (Bos indicus) and their crosses with Bos taurus. Meat. Sci. 123, 64-66. doi 10.1016/j.meatsci.2016.09.003 [ Links ]
Fan, S.J., Li, H.B., Cui, G., Kong, X.L., Sun, L.L., Zhao, Y.Q., Li, Y.H. & Zhou, J., 2016. miRNA-149* promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia. Leuk. Res. 41, 62-70. doi 10.1016/j.leukres.2015.11.016 [ Links ]
Han, R., Wei, Y., Kang, X., Chen, H., Sun, G., Li, G., Bai, Y., Tian, Y. & Huang, Y., 2012. Novel SNPs in the PRDM16 gene and their associations with performance traits in chickens. Mol. Biol. Rep. 39, 3153-3160. doi 10.1007/s 11033-011-1081-y [ Links ]
Imbar, T. & I. Eisenberg, 2014. Regulatory role of microRNAs in ovarian function. Fertil. Steril. 101, 1524-1530. doi 10.1016/j.fertnstert.2014.04.024 [ Links ]
Jazdzewski, K., Murray, E.L., Franssila, K., Jarzab, B., Schoenberg, D.R. & de la Chapelle, A., 2008. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA. 105(20), 7269-7274. doi:10.1073/pnas.0802682105 [ Links ]
Juengel, J.L., Mosaad, E.M.O., Mitchell, M.D., Phyn, C.V.C., French, M.C., Meenken, E.D., Burke, C.R. & Meier, S., 2022. Relationships between prostaglandin concentrations, SNP in HSD17B12, and reproductive performance in dairy cows. J. Dairy. Sci. doi 10.3168/jds.2021-21298 [ Links ]
Kabekkodu, S.P., Shukla, V., Varghese, V.K., Chakrabarty, D.S.J.S. & Satyamoorthy, K., 2018. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 93, 1955-1986. doi 10.1111/brv.12428 [ Links ]
Kong, Y., Yuan, Z., Liu, X., Li, F. & Yue, X., 2022. A novel SNP within LIPE gene is highly associated with sheep intramuscular fat content. Small. Ruminant. Res. 209. doi 10.1016/j.smallrumres.2022.106658 [ Links ]
Kotaki, R., Koyama-Nasu, R., Yamakawa, N. & Kotani, A., 2017. miRNAs in normal and malignant hematopoiesis. Int. J. Mol. Sci. 18(7), 1495. doi:10.3390/ijms18071495 [ Links ]
Kumar, S., Singh, U., Deb, R., Tyagi, S., Mandal, D.K., Kumar, M., Sengar, G., Sharma, S., Singh, R. & Singh, R., 2015. A SNP (g.358A>T) at intronic region of CD9 molecule of crossbred bulls may associate with spermatozoal motility. Meta. Gene. 5, 140-143. doi 10.1016/j.mgene.2015.07.004 [ Links ]
Lazare, S.S., Wojtowicz, E.E., Bystrykh, L.V. & de Haan, G., 2014. microRNAs in hematopoiesis. Exp. Cell. Res. 329, 234-238. doi 10.1016/j.yexcr.2014.08.033 [ Links ]
Li, H., Sun, G.R., S. Lv, J., Wei, Y., Han, R.L., Tian, Y.D. & Kang, X.T., 2012. Association study of polymorphisms inside the miR-1657 seed region with chicken growth and meat traits. Br. Poult. Sci. 53, 770-776. doi 10.1080/00071668.2012.750716 [ Links ]
Li, H., Tian, Y., Sun, G., Liu, X., Jiang, R., Han, R., Li, G. & Kang, X., 2014. Association study of a common genetic variant in pre-miR-1596 with chicken performance traits. Mol. Biol. Rep. 41, 7175-7181. doi 10.1007/s11033-014-3600-0 [ Links ]
Li, H., Wang, S., Yan, F., Liu, X., Jiang, R., Han, R., Li, Z., Li, G., Tian, Y., Kang, X. & Sun, G., 2015. Effect of polymorphism within miRNA-1606 gene on growth and carcass traits in chicken. Gene. 566, 8-12. doi 10.1016/j.gene.2015.03.037 [ Links ]
Luinenburg, D.G. & de Haan, G., 2020. MicroRNAs in hematopoietic stem cell aging. Mech. Ageing. Dev. 189, 111281. doi 10.1016/j.mad.2020.111281 [ Links ]
Maalouf, S.W., Liu, W.S. & Pate, J.L., 2016. MicroRNA in ovarian function. Cell. Tissue. Res. 363, 7-18. doi 10.1007/s00441-015-2307-4 [ Links ]
Martin, E.C., Qureshi, A.T., Dasa, V., Freitas, M.A., Gimble, J.M. & Davis, T.A., 2016. MicroRNA regulation of stem cell differentiation and diseases of the bone and adipose tissue: Perspectives on miRNA biogenesis and cellular transcriptome. Biochimie. 124, 98-111. doi 10.1016/j.biochi.2015.02.012 [ Links ]
Mehta, A., Zhao, J.L., Sinha, N., Marinov, G.K., Mann, M., Kowalczyk, M.S., Galimidi, R.P., Du, X., Erikci, E., Regev, A., Chowdhury, K. & Baltimore, D., 2015. The microRNA-132 and microRNA-212 cluster regulates hematopoietic stem cell maintenance and survival with age by buffering FOXO3 expression. Immunity. 42, 1021-1032. doi 10.1016/j.immuni.2015.05.017 [ Links ]
Mencia, A., Modamio-Hoybjor, S., Redshaw, N., Morin, M., Mayo-Merino, F., Olavarrieta, L., Aguirre, L.A., del Castillo, I., Steel, K.P., Dalmay, T., Moreno, F. & Moreno-Pelayo, M.A., 2009. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 09-613. doi 10.1038/ng.355 [ Links ]
Momoi, Y., Tsusaki, T., Yamashita, H. & Takahashi, H., 2021. The effectiveness of an SNP marker in the cholecystokinin type A receptor gene for improving growth traits in Amakusa Daioh cross chickens. J. Appl. Poultry. Res. 30. doi 10.1016/j.japr.2021.100166 [ Links ]
Qiang, J., Tao, Y.F., Bao, J.W., Chen, J., Li, H.X., He, J. & Xu, P., 2018. High fat diet-induced miR-122 regulates lipid metabolism and fat deposition in genetically improved farmed tilapia (GIFT, Oreochromis niíoticus) Liver. Front. Physiol. 9, 1422. doi 10.3389/fphys.2018.01422 [ Links ]
Rani, V. & Sengar, R.S., 2022. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 119, 685-692. doi 10.1002/bit.28029 [ Links ]
Rasko, J.E. & Wong, J.J., 2017. Nuclear microRNAs in normal hemopoiesis and cancer. J. Hematol. Oncol. 10(1), 8. doi 10.1186/s13045-016-0375-x [ Links ]
Salim, U., Kumar, A., Kulshreshtha, R. & Vivekanandan, P., 2022. Biogenesis, characterization, and functions of mirtrons. WIRES. RNA. 13(1), e1680. doi 10.1002/wrna.1680 [ Links ]
Sand, M., Gambichler, T., Sand, D., Skrygan, M., Altmeyer, P. & Bechara, F.G., 2009. MicroRNAs and the skin: Tiny players in the body's largest organ. J. Dermatol. Sci. 53(3), 169-175. doi 10.1016/j.jdermsci.2008.10.004 [ Links ]
Shi, J. & Sun, G., 2017. Effect of pre-miRNA-1658 gene polymorphism on chicken growth and carcass traits. Asian- Australas. J. Anim. Sci. 30(4), 455-461. doi 10.5713/ajas.16.0305 [ Links ]
Song, C., Yao, J., Cao, C., Liang, X., Huang, J., Han, Z., Zhang, Y., Qin, G., Tao, C., Li, C., Yang, H., Zhao, J., Li, K. & Wang, Y. 2016. PPARgamma is regulated by miR-27b-3p negatively and plays an important role in porcine oocyte maturation. Biochem. Biophys. Res. Commun. 479(2), 224-230. doi 10.1016/j.bbrc.2016.09.046 [ Links ]
Vienberg, S., Geiger, J., Madsen, S. & Dalgaard, L.T., 2017. MicroRNAs in metabolism. Acta. Physiol. (Oxf). 219, 346-361. doi 10.1111/apha.12681 [ Links ]
Wang, J., Yang, L.Z., Zhang, J.S., Gong, J.X., Wang, Y.H., Zhang, C.L., Chen, H. & Fang, X.T., 2018. Effects of microRNAs on skeletal muscle development. Gene. 668, 107-113. doi 10.1016/j.gene.2018.05.039 [ Links ]
Wang, Q., Wang, Q., Melak, S., Lin, X., Wei, W., Zhang, L. & Chen, J., 2021. A novel c.-652C>T mutation in UCHL1 gene is associated with the growth performance in Yangzhou goose. Poult. Sci. 100(7), 101089. doi 10.1016/j.psj.2021.101089 [ Links ]
Wang, X., Gu, Z. & Jiang, H., 2013. MicroRNAs in farm animals. Animal. 7(10), 1567-1575. doi 10.1017/S1751731113001183 [ Links ]
Wang, X., Sun, S., Tong, X., Ma, Q., Di, H., Fu, T., Sun, Z., Cai, Y., Fan, W., Wu, Q., Li, Y., Wang, Q. & Wang, J., 2017. MiRNA-154-5p inhibits cell proliferation and metastasis by targeting PIWIL1 in glioblastoma. Brain. Res. 1676, 69-76. doi 10.1016/j.brainres.2017.08.014 [ Links ]
Wang Y, Xu, H.Y., Gilbert, E.R., Peng, X., Zhao, X.L., Liu, Y.P. & Zhu, Q. 2014. Detection of SNPs in the TBC1D1 gene and their association with carcass traits in chicken. Gene. 547(2), 288-294. doi 10.1016/j.gene.2014.06.061 [ Links ]
Wu, P.X., Zhou, J., Wang, K., Chen, D.J., Yang, X.D., Liu, Y.H., Jiang, A.A., Shen, L.Y., Jin, L., Xiao, W.H., Jiang, Y.Z., Li, M.Z., Zhu, L., Zeng, Y.S., Xu, X., Qiu, X.T., Li, X.W. & Tang, G.Q., 2021. Identifying SNPs associated with birth weight and days to 100 kg traits in Yorkshire pigs based on genotyping-by-sequencing. J. Integr. AGR. 20(9), 2483-2490. doi 10.1016/s2095-3119(20)63474-8 [ Links ]
Yang, Q., Wu, P., Wang, K., Chen, D., Zhou, J., Ma, J., Li, M., Xiao, W., Jiang, A., Jiang, Y., Bai, L., Zhu, L., Li, X. & Tang, G., 2019. SNPs associated with body weight and backfat thickness in two pig breeds identified by a genome-wide association study. Genomics. 111(6), 1583-1589. doi 10.1016/j.ygeno.2018.11.002 [ Links ]
Yuan, S., Schuster, A., Tang, C., Yu, T., Ortogero, N., Bao, J., Zheng, H. & Yan, W., 2016. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 143(4), 635-647. doi 10.1242/dev.131755 [ Links ]
Zerehdaran, S., Vereijken, A.L., van Arendonk, J.A. & van der Waaijt, E.H., 2004. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult. Sci. 83(4), 521-525. doi 10.1093/ps/83.4.521 [ Links ]
Zhang, D., Wang, Y., Ji, Z. & Wang, Z., 2016. Identification and differential expression of microRNAs associated with fat deposition in the liver of Wistar rats with nonalcoholic fatty liver disease. Gene. 585(1), 1-8. doi 10.1016/j.gene.2016.03.011 [ Links ]
Zhang, J., Guan, Y., Shen, C., Zhang, L. & Wang, X., 2019a. MicroRNA-375 regulates oocyte in vitro maturation by targeting ADAMTS1 and PGR in bovine cumulus cells. Biomed. Pharmacother. 118, 109350. doi 10.1016/j.biopha.2019.109350 [ Links ]
Zhang, J., Xu, Y., Liu, H. & Pan, Z., 2019b. MicroRNAs in ovarian follicular atresia and granulosa cell apoptosis. Reprod. Biol. Endocrinol. 17(1), 9. doi 10.1186/s12958-018-0450-y [ Links ]
Zhao, F., Liu, X., Wang, Z., Lang, H., Zhang, T., Wang, R., Lin, X., He, D., Shi, P. & Pang, X., 2020. Novel mouse miRNA, Chr13_novelMiR7354-5p, improves bone-marrow-derived mesenchymal stem cell differentiation into insulin-producing cells. Mol. Ther. Nucleic. Acids. 19, 1110-1122. doi 10.1016/j.omtn.2020.01.001 [ Links ]
Submitted 25 October 2022
Accepted 12 April 2023
Published 12 July 2023
# Corresponding author: G.R. Sun 20122031@nynu.edu.cn; L.G. Yao lunguangyao@163.com
ORCID: J.Z. Shi https://orcid.org/0000-0002-6423-7248; Y.W. Wang https://orcid.org/0000-0001-6429-3154; T.J. Wang https://orcid.org/0000-0003-2944-2040; J. Ji https://orcid.org/0000-0001-9511-4163; G.R. Sun https://orcid.org/0000-0002-2141-913X; L.G. Yao https://orcid.org/0000-0003-4059-1100














