Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.53 n.3 Pretoria 2023
http://dx.doi.org/10.4314/sajas.v53i3.08
Effects of diet on ruminal methanogenic archaea composition and diversity in cashmere goats
K.N. Li Y.R. Wei; R.H. Na#
College of Animal Science, Inner Mongolia Agricultural University, 306 Zhaowuda Road, Hohhot 010018, Inner Mongolia, China
ABSTRACT
Diet is the most direct way to affect the rumen microbial community. The objective of this study was to examine the effects of different diets on ruminal methanogen composition in cashmere goats. Twenty-four cashmere goats were randomly divided into four groups that were fed alfalfa hay (AH), alfalfa hay plus concentrate (AHC), corn stalks (CS), and corn stalks plus concentrate (CSC). The concentration of ammonia-N for the group fed AHC was substantially higher than in the other groups. The concentrations of total VFAs, acetate, and propionate in AH and AHC groups were higher than in the CS and CSC groups. The abundance of archaea was markedly different among different groups based on a high-throughput sequencing method. The abundance of Cand;'datus_Methanomethylophilus was 21.28% in the AH group, which was higher than the other groups. (Unclassified_c_ Thermoplasmata was the dominant methanogen in the AH and AHC groups, with abundances of 48.09% and 44.97%, respectively; Methanobrevibacter was the dominant methanogen in the CS and CSC groups, with abundances of 41.22% and 20.73%, respectively. A positive correlation was identified between pH and Methanosphaera; a negative relationship was observed between acetate and (Unclassified_o_Thermoplasmatales. In conclusion, the methanogen communities in cashmere goats varied on different diets. In the alfalfa hay-based diets, the Methanomassiliicoccales-affiliated groups were the dominant methanogens in the rumen of the goats. However, when fed the corn stalk-based diets, Methanobrev;bacter was the dominant methanogen in the rumen fluid. The results were related to dietary composition, especially crude protein content.
Keywords: forage, methane production, methanogens, rumen fermentation
Introduction
Ruminants provide high-quality protein for humans, such as beef, mutton, and dairy products. The rumen of ruminants has the unique ability to utilize various feeds, which is mainly attributed to the complex, diverse, nonpathogenic microorganisms inhabiting the rumen, including bacteria, fungi, protozoa, and archaea (Kittelmann et al., 2013). Methanogens are a phylogenetically diverse group of archaea. The function of methanogens is to produce methane via the hydrogenotrophic pathway. This process keeps a low H2 pressure in the rumen to promote the digestion of various raw materials by fungi, protozoa, and bacteria (Moss et al., 2000). To a much lesser extent, methane can also be produced in the rumen through the utilization of methyl groups (methylotrophic pathway) and less commonly from acetate (Leahy et al., 2010). The most studied methanogenesis pathway in the rumen is the hydrogenotrophic pathway, which accounts for >90% of the total methane production from the rumen. However, methane cannot be used by ruminants and is emitted into the atmosphere through eructation. This accounts for approximately 6% of global anthropogenic greenhouse gas emissions (Gerber et al., 2013). Furthermore, enteric methane emissions are an important energy loss in the process of ruminant livestock feeding, accounting for 2-12% of the gross energy consumed by ruminants (Johnson & Johnson, 1995).
Enteric methane production is affected by many factors, including diet, feed intake, the digestibility of the diet in the rumen, the rumen passage rate of the diet, rumen pH, and animal genotype and phenotype (Beauchemin et al., 2020). Previous studies on beef cattle, dairy cows, goats, and sheep have shown that dietary manipulation can affect the methanogen community composition and is an effective way to reduce enteric methane emissions in ruminant production systems (Yang et al., 2012; Pedreira et al., 2013; Philippeau et al., 2017; Lima et al., 2019).
Forage type is recognized to be an important factor in the composition of microbial communities, rumen fermentation, and rumination time in ruminants (Liu et al., 2016). One type of forage, corn stalks, is abundant and low-cost but is typically considered poor-quality roughage. A second type, alfalfa, is well known for its high quality and plays an essential part in the production of ruminants. Due to differences in the physical structure and chemical properties, alfalfa and corn stalks in the rumen produce different emissions of enteric methane. However, the difference in the composition of colonized ruminal methanogen communities of the two forages may also be an important factor. Cashmere goats are an important ruminant species in many regions of the world, including China, Russia, India, and Mongolia. However, little is known about the effect of alfalfa hay- or corn stalk-based diets on the composition and diversity of methanogenic archaea in cashmere goats.
A range of molecular technologies have been used to determine the diversity and phylogeny of methanogens. Unlike the traditional method involving Sanger sequencing of 16S rRNA, the methyl coenzyme M reductase (mcrA) gene is an alternative strategy to detect the presence, abundance, and activity of methanogens under various conditions (Tomkins et al., 2015). The mcrA gene is highly conserved phylogenetically and encodes the methyl coenzyme, M reductase, which catalyses the terminal step of methanogenesis in all methanogenic archaea (Friedrich 2005). Sirohi et al. (2013) reported that diversity analysis using the mcrA gene provides better insight into ruminal methanogens than the 16S rRNA gene. Therefore, the objective of the present study was to examine the effects of alfalfa hay- and corn stalk-based diets on fermentation profile and the composition and diversity of methanogens in rumen fluid of cashmere goats.
Materials and methods
All animal procedures were conducted following the 'Laboratory Animal Guideline for Ethical Review of Animal Welfare,' National Standard of the People's Republic of China (GB/T 35892-2018). The care and use of animals fully complied with local animal welfare laws, guidelines, and policies.
Twenty-four healthy, 8-month-old cashmere ewes (average body weight 25.2 ± 1.8 kg) were selected and randomly divided into four treatments consisting of six replicates with one goat per replicate. The dietary treatments were as follows: alfalfa hay-fed group (AH), alfalfa hay plus concentrate-fed group (AHC), corn stalk-fed group (CS), and corn stalk plus concentrate-fed group (CSC). The ratio of concentrate to forage in AHC and CSC groups was 25:75. Animals were individually fed twice a day, at 08:00 and 17:00. All animals had ad libitum access to water. The Feed Research Institute of the Chinese Academy of Agricultural Sciences was entrusted with the analysis of conventional indices of feeds. The compositions and nutrient levels of the diets are shown in Table 1. Before the start of the experiment, the goats were dewormed, and the animal shed was fully disinfected. The experiment lasted for 40 days, with a 20-day adaptation period and a 20-day formal experimental period.
On day 20 of the formal experimental period, an oral stomach tube was used to collect ruminal fluid from the 24 animals for analysis of ruminal fermentation parameters and diversity of methanogens.
For each animal, 100 mL of ruminal fluid was collected and the initial 50 mL was discarded to avoid contamination by saliva; the last 50 mL of ruminal fluid was retained. The ruminal fluid was filtered with four layers of cheesecloth, and 2 mL of the strained ruminal filtrate was taken to measure the pH value immediately, using a portable pH meter (Starter 300; Ohaus Instruments Co. Ltd., Shanghai, China). A total of 4 mL of the filtrate was aliquoted into cryogenic vials, quickly frozen in liquid nitrogen, and stored at -80 °C until microbial DNA extraction. The concentrations of VFAs were determined using a previously reported method (Hoskin et al., 1995). An amount of 10 mL of the filtrate was centrifuged at 12 000 x g at 4 °C for 5 min, and the supernatant from each sample was then transferred to 2 mL tube. After adding 25% (w/v) metaphosphoric acid solution to the fluid at a volume ratio of 1:5, the mixture was vortexed for 10 s and centrifuged at 2 000 x g for 20 min to remove the precipitate. The supernatant was then applied to a GC-8A gas chromatograph (Shimadzu Corp., Kyoto, Japan) to measure the concentrations of the main VFAs. A 5 mL sample of each ruminal filtrate sample was centrifuged at 3 500 x g for 10 min, and the supernatants were used to determine the ammonia-N (NH3-N) concentrations using the methods described by Chaney & Marbach (1962).
Microbial DNA was extracted from rumen fluid samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, CA, USA). The integrity of the extracted DNA was detected using 1% agarose gel electrophoresis; the concentration and purity of the extracted DNA were determined using a NanoDrop 2000 UV-vis (ultraviolet-visible) spectrophotometer (Thermo Scientific, Wilmington, NC, USA). DNA samples that met the requirements were subjected to MiSeq sequencing. The mcrA-specific primers used in the present study were MLrF (5'-ACTCCTACGGGAGGCAGCA-3') and MLrR (5'-GGACTACHVGGGTWTCTAAT-3'). The polymerase chain reaction (PCR) reaction volume was 20 μL, including 10 ng of template DNA, 0.8 μL of each primer (forward and reverse) with a concentration of 5 μmol L-1, 4 μL of 5x FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.4 μL of FastPfu polymerase, 0.2 μL of bovine serum albumin, and ddH2O. The PCR protocol was 95 °C for 3 min; 27 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s; then 72 °C for 10 min. The PCR products were subjected to gel purification using an AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, USA), and quantified using QuantiFluor™-ST (Promega, Fitchburg, WI, USA). The purified amplicons were pooled on an Illumina MiSeq PE300 instrument (Illumina, San Diego, CA, USA) and were equimolar and paired-end sequenced (2 x 300) according to the standard protocols by Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China).
The raw FASTQ files were demultiplexed and quality-filtered using QIIME (version 1.9.1) using the following criteria: (i) the 300-bp reads were truncated at any site that had an average quality score <20 over a 50-bp sliding window, and truncated reads <50 bp were discarded; (ii) the paired-end reads were merged into a sequence with a minimum overlap length of 10 bp; (iii) the allowed maximum mismatch ratio of the overlap region of the spliced sequence was 0.2, and the nonconforming sequence was screened; and (iv) samples were distinguished according to the barcode and primer at the head and tail ends of the sequence, and the sequence direction was adjusted. The allowable number of barcode mismatches was 0, and the maximum number of primer mismatches was 2.
UPARSE software (Highly Accurate OTU Sequences from Microbial Amplicon Reads, version 7.0.1090, http://www.drive5.com/uparse/) was used to cluster the sequences into operational taxonomie units (OTUs; based on similarity >97%) and chimeric sequences were identified and removed using UCHIME (Chimera Prediction for Amplicon Sequencing). OTUs were used for alpha diversity (Coverage, Sobs, Ace, Chao, Shannon, and Simpson) analysis. OTUs were taxonomically analysed using the Ribosomal Database Project (RDP) Classifier algorithm (version 2.11 http://sourceforge.net/projects/rdp-classifier/) against the FunGene Database, using a confidence threshold of 70% (Amato et al., 2013). The rarefaction curve analysis using Mothur v.1.21.1 was performed to reflect the sequencing depth. The composition of the methanogen community was compared by principal coordinate analysis (PCoA) using the Bray-Curtis dissimilarity metric, with sequence identity >97% at the OTU level to perform visual calculation and analysis of the beta diversity of samples.
The data ruminal fermentation parameters were analysed in a completely randomized design using the Proc Mixed procedure of SAS (SAS Inst. Inc., Cary, NC, USA). According to this statistical model:
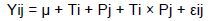
where: Yij was the dependent, continuous variable; μ was the overall mean; Ti was the fixed effect of diet treatment (i = alfalfa hay, alfalfa hay plus concentrate, corn stalk, and corn stalk plus concentrate); Pj was the fixed effect of the pen (j = 1, 2, 3, 4, 5, and 6); Ti x Pj was the fixed effect of the interaction between diet and pen, and cij was the residual error. The mixed model included fixed effects of diet and random effects of the pen. The pen was considered the experimental unit and the repeated measurement. Data means significance was declared at P <0.05.
For evaluation of ruminal methanogen diversity indices and the methanogen community structure at phylum and genus level, a one-way analysis of variance (ANOVA) and Duncan's multiple range tests were carried out in SAS (SAS Inst. Inc., Cary, NC, USA). Relative abundance data are presented as percentages, and the correlation between fermentation parameters and methanogens was examined using Spearman's correlations.
Results
The effects of diet on ruminal fermentation parameters are shown in Table 2. The concentration of NH3-N in group AHC was higher than in the other groups (P <0.01). The concentrations of total VFAs in group AH and AHC were higher than in group CS (P <0.05). The acetate concentration in group AH and AHC was higher than that in group CSC (P <0.05), and the concentration of propionate in group AH and AHC was higher than that in group CS and CSC (P <0.01). The differences in other ruminal fermentation parameters were not significant (P >0.05).
After sequence processing, a total of 410 247 high-quality sequences were obtained. The number of sequences for each sample ranged from 11 488 to 23 455. Rarefaction curves of methanogens based on the Shannon index at the OTU level are shown in Figure 1. The curves tended to be flat, indicating that the amount of sequencing data was large enough to reflect the diversity of the methanogens in the sample. Table 3 shows the richness and diversity indices of methanogens in the rumen of cashmere goats fed different diets. The Good's coverage of each group was >0.99, indicating that the sampling and sequencing reflected the diversity of the methanogens that were present. The Simpson index in group AHC was higher than that in group AH and CS (P < 0.05). However, the differences in the Sobs, Shannon, Ace, or Chao indices were not significant (P >0.05). A Venn diagram was constructed at the OTU level based on the sequences obtained from ruminal fluid samples from cashmere goats (Figure 2). A total of 106 OTUs were obtained from all samples, of which 47 OTUs existed in all groups (defined as core OTUs). The core OTUs comprised approximately 44.34% of the total OTUs. In addition, 10, 5, 4, and 3 OTUs were uniquely identified in groups AH, AHC, CS, and CSC, respectively. The ordination plot showed apparent spatial separation among the AH, AHC, CS, and CSC group based on methanogen communities (Figure 3).
The relative abundance of the various methanogenic archaea identified using the mcrA gene sequencing approach is shown in Table 4. Taxonomy-based analysis revealed that two phyla and six genera were detected across the 24 samples. At the phylum level, the relative abundance of Euryarchaeota in group AH and CS was higher than in group CSC (P <0.05). The relative abundance of Unclassified_d_Unclassified in group CSC (33.09%) was higher than that in group AH (0.70%) (P <0.05) and CS (8.60%) (P <0.05). At the genus level, the relative abundance of Candidatus_Methanomethylophilus in group AH (21.28%) was higher than group AHC (6.92%), CS (4.00%), and CSC (3.49%) (P <0.05).
The correlation heatmap was based on Spearman's correlation coefficient for the relationship between the methanogenic archaea community (at the genus level) and ruminal fermentation variables (Figure 4). The pH value was positively (P <0.05) related to the presence of Methanosphaera. A negative relationship (P <0.05) was observed between acetate and Unclassified_o_The rmoplasmatales.
Discussion
Diets are substrates for rumen microbial fermentation; their nutritional composition and physical properties directly affect rumen fermentation patterns in ruminants. NH3-N is an important parameter to reflect the rumen fermentation index. The concentrations of ammonia in the rumen represent a balance between formation (by microbial degradation of protein, amino acids, urea, and nucleic acids) and removal (by synthesis of microbial biomass, absorption across the rumen epithelium, and by outflow to the omasum) (Knight et al., 2011). In the current study, the concentration of NH3-N in group AHC was substantially higher than in groups AH, CS, and CSC. In addition, the concentrations of the propionate, acetate, and total VFAs were higher in group AH and AHC than in group CS and CSC. These results may be related the higher readily degradable fraction of carbohydrates and protein in alfalfa (Wang et al., 2019). Compared with corn stalks, alfalfa hay contains a higher proportion of nonfibrous carbohydrates and a lower proportion of neutral detergent fibre (NDF). In this study, the NDF content in group AH (57.76%) was lower than in group CS (71.32%). Mccaughey et al. (1999) studied the effects of different types of forage on beef cattle methane production and found that, compared with pure gramineous forage grass, mixed forage of alfalfa and gramineous forage grass had a lower fibre content and higher rumen flow rate, and the methane production was reduced by 9%. When the proportion of NDF in dairy cow diets was increased from 31.5% to 38%, the daily methane production increased by 23% (Dong et al., 2019). Therefore, reducing the content of NDF is an effective method to reduce methane production from ruminants. According to previous reports, methane production from concentrate-based diets is lower than that from forage-based diets (Pedreira et al., 2013). The reason is that fermentation of the starch in the rumen will produce more propionate, and the increased level of propionate can compete with methanogens for H2, thereby reducing methane production. Furthermore, starch has a faster digestion and fermentation rate than cellulose, which will lower the rumen pH, thereby inhibiting the growth of methanogens (Beauchemin et al., 2020). In the present work, because of the small proportion of concentrate added to the diets, the rumen pH value in groups AH, AHC, CS, and CSC did not differ substantially.
The diet, as a substrate for microbial fermentation, its physical properties, and nutritional composition directly affects methanogenic archaea diversity. In this experiment, by using a high-throughput sequencing method based on the mcrA gene, we investigated the α-diversity and β-diversity indices of ruminal methanogens in cashmere goats on different diets. The Simpson index observed in this study was substantially influenced by the diet, and the PCoA chart showed apparent spatial separation among the AH, AHC, CS, and CSC groups based on methanogen communities. These data indicate that the different diets had a marked effect on the composition of methanogen communities in the rumen. In previous studies, similar results were observed by Franzolin et al. (2012), when analyzing the ruminal methanogenic archaea communities of water buffalo fed different diets. The different diets had a marked effect on the ruminal methanogen population structure. Dong et al. (2019) performed high-throughput sequencing based on mcrA and found that changes in the liquid diet, solid diet, and weaning time of Holstein's calves led to changes in the composition of the methanogens in the rumen.
Typically, archaeal communities account for only 3-4% of the rumen microbiome, while Euryarchaeota are the dominant archaea in the rumen (Kumar et al., 2015). Wang et al. (2017) reported that Euryarchaeota accounted for 82% of the composition of methanogens in the ruminal fluid of goats, which is in accordance with the methanogen densities found in this study. In the present study, the mean relative abundance of Euryarchaeota accounted for approximately 84% of the total sequences. In addition, the relative abundance of Euryarchaeota in group AH was higher than in groups CS and CSC. Shi et al. (2014) and Danielsson et al. (2017) reported weak or no relationships between the methane production of individual animals and the total abundance of archaea in their rumen. By contrast, it seems that the composition of the methanogenic archaeal community is highly correlated with methane production. Wallace et al. (2015) applied metagenomics on the rumen microbial community of dairy cows and found that the relative abundance of Methanobrevibacter in the rumen of dairy cows with high methane production was higher than that of dairy cows with low methane production. In the present study, the dominant methanogenic archaeal identified in the rumen of the cashmere goats varied among the different treatments. (Unclassified_c_Thermoplasmata was the overwhelming dominant methanogen in group AH and AHC at the genus level, whereas Methanobrevibacter was the overwhelming dominant methanogen in group CS and CSC. Therefore, we conclude that the differences in methane emissions between diets with corn stalk as roughage and those with alfalfa hay as roughage may be related to Methanobrevibacter. Moreover, the relative abundance of (Unclassified_c_Thermoplasmata was highest in groups AH and AHC. This may be caused by the variable production of methylated substrate compounds (methylamine and methanol) in the rumen, which is mediated by alfalfa hay. Peng et al. (2021) reported that the methanol-utilizing archaea, Thermoplasmata, was enriched on alfalfa, which had the highest pectin content of all the four substrates they used (alfalfa stems, bagasse, reed canary grass, and xylan). A primary degradation product of pectin is methanol and this probably explains the enrichment of Thermoplasmata on alfalfa.
In the present study, the acetate concentration was negatively related to (Unclassified_o_Thermoplasmatales, possibly because Thermoplasmatales reduce methanol or other methylated compounds for methanogenesis. Extensive evidence has demonstrated that the formation of acetate results in the production of additional methanogenic substrates (formate and H2) (Danielsson et al., 2017). Most previous studies that have investigated the structure of the methanogen community have found that Methanobrevibacter phylotypes are the dominant methanogens in the rumen of different ruminants worldwide (Fouts et al., 2012; Seedorf et al., 2015). However, less research has reported that the archaeal community is dominated by the methanogenic archaea variably classified as the members from the seventh order of Thermoplasmatales, i.e., Methanomassiliicoccales (previously called Rumen Cluster C, RCC) (Borrel et al., 2014; Noel et al., 2016). Iino et al. (2015) reported that the Methanomassiliicoccales mainly produce methane through the methylotrophic pathway and can use methylamine and methanol as substrates for methanogenesis. Methanomassiliicoccales belong to a branch of Thermoplasmata (Tajima et al., 2001). (Unclassified_c_Thermoplasmata and Candidatus_Methanomethylophilus belong to the Methanomassiliicoccales-affiliated groups (Noel et al., 2016). The present study showed that (Unclassified_c_Thermoplasmata, Methanobrevibacter, Methanosphaera, and Candidatus_Methanomethylophilus were the dominant methanogens in the rumen fluid of cashmere goats. This result concurs with other studies on the composition of rumen methanogen communities in ruminants (Wang et al., 2017; Zhu et al., 2017). The current results showed that the relative abundance of Candi'datus_Methanomethylophilus was higher in group AH than in the other groups, and Unclassifi'ed_c_Thermoplasmata was the overwhelming dominant methanogen in group AH and AHC. We speculate that the differences in methane production in group AH and group CS may be related to the relative abundance of Methanomassiliicoccales-affiliated groups, but further studies are needed to confirm this. In the present study, we also found the Methanomassiliicoccales-affiliated groups were the dominant methanogens found in the alfalfa hay-based diets, and Methanobrevibacter was the main methanogen found in the corn stalk-based diets. These results may be related to the chemical composition of the diet. Compared with the alfalfa hay-based diets, the corn stalk-based diets had lower crude protein content. However, there are few reports on the effects of dietary crude protein on the methanogenic archaea community. Furthermore, in the present study, the pH value was positively related to Methanosphaera. The rumen pH is a consequence of feed degradation and microbial function. Dong et al. (2019) also reported the pH value was substantially related to the presence of Methanosphaera in the rumen of Holstein bull calves. This concurs with the current study. However, further studies should be conducted to explore the relationship between pH value and Methanosphaera.
Conclusion
In summary, the ruminal fermentation parameters and the methanogen communities in the rumen of cashmere goats varied when fed different diets. With the alfalfa hay-based diets, the Methanomassiliicoccales-affiliated groups were the dominant methanogen found in the rumen fluid of the goats. However, when fed the corn stalk-based diets, Methanobrev;bacter was the main methanogen found in the rumen fluid. These results are related to dietary composition, especially crude protein content. These analyses provide further insight into diets that can be used to mitigate enteric methane emissions in ruminant livestock production systems.
Acknowledgments
The authors are grateful for the financial support provided by the National Natural Science Foundation of China (No. 31760690), and the Program for Innovative Research Team in Universities of Inner Mongolia Autonomous Region (NMGIRT2322). The author thanks his laboratory colleagues for their assistance in the data and sample collection and laboratory analysis.
Authors' Contributions
KNL was responsible for laboratory analyses, data collection, analysis of results, and writing the manuscript. YRW contributed to laboratory analyses and data acquisition. RHN supervised the project planning and statistical analysis of the results and critically reviewed the manuscript.
Conflict of interest declaration
The authors declare there are no competing interests.
References
Beauchemin, K.A., Ungerfeld, E.M., Eckard, R.J. & Wang, M., 2020. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal. 14(1), s2-s16. Doi: 10.1017/S1751731119003100. [ Links ]
Borrel, G., Parisot, N., Harris, H.M., Peyretaillade, E., Gaci, N., Tottey, W., Bardot, O., Raymann, K., Gribaldo, S., Peyret, P., O'Toole, P.W. & Brugère, J.F., 2014. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics. 15(1), 679-702. Doi: 10.1186/1471-2164-15-679. [ Links ]
Cammell, S.B., Thomson, D.J., Beever, D.E., Haines, M.J., Dhanoa, M.S. & Spooner, M.C., 1986. The efficiency of energy utilization in growing cattle consuming fresh perennial ryegrass (Lolium perenne cv. Melle) or white clover (Trifolium repens cv. Blanca). Brit. J. Nutr. 55, 669-680. Doi: 10.1079/BJN19860073. [ Links ]
Chaney, A.L. & Marbach, E.P., 1962. Modified reagents for determination of urea and ammonia. Clin. Chem. 8(2), 130-132. Doi: 10.1093/clinchem/8.2.130. [ Links ]
Danielsson, R., Dicksved, J., Sun, L., Gonda, H., Müller, B., Schnürer, A. & Bertilsson, J., 2017. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 8, 226-240. Doi: 10.3389/fmicb.2017.00226. [ Links ]
Dong, L.F., Li, B.C. & Diao, Q.Y., 2019. Effects of dietary forage proportion on feed intake, growth performance, nutrient digestibility, and enteric methane emissions of Holstein heifers at various growth stages. Animals. 9(10), 725-738. Doi: 10.3390/ani9100725. [ Links ]
Dong, L.F., Ma, J.N., Tu, Y. & Diao, Q.Y., 2019. Weaning methods affect ruminal methanogenic archaea composition and diversity in Holstein calves. J. Integr. Agr. 18(5), 1080-1092. Doi: 10.1016/s2095-3119(18)62120-3. [ Links ]
Fouts, D.E., Szpakowski, S., Purushe, J., Torralba, M., Waterman, R.C., MacNeil, M.D., Alexander, L.J. & Nelson, K.E., 2012. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS ONE. 7(11), e48289. Doi: 10.1371/journal.pone.0048289. [ Links ]
Franzolin, R., St-Pierre, B., Northwood, K. & Wright, A.D., 2012. Analysis of rumen methanogen diversity in water buffaloes (Bubalus bubalis) under three different diets. Microb. Ecol. 64(1), 131-139. Doi: 10.1007/s00248-012-0007-0. [ Links ]
Friedrich, M.W., 2005. Methyl-coenzyme M reductase genes: Unique functional markers for methanogenic and anaerobic methane-oxidizing archaea. Method. Enzymol. 397, 428-442. Doi: 10.1016/s0076-6879(05)97026-2. [ Links ]
Gerber, P.J., Steinfeld, H., Henderson, B., Mottet, A., Opio, C., Dijkman, J. & Tempio, G., 2013. Tackling climate change through livestock: A global assessment of emissions and mitigation opportunities. Food and Agriculture Organization of the United Nations (FAO). Rome. Doi: 10.1109/IPECON.2010.5697145. [ Links ]
Hoskin, S.O., Stafford, K.J. & Barry, T.N., 1995. Digestion, rumen fermentation, and chewing behaviour of red deer fed fresh chicory and perennial ryegrass. J. Agr. Sci. 124(2), 289-295. Doi: 10.1017/s0021859600072956. [ Links ]
Iino, T., Tamaki, H., Tamazawa, S., Ueno, Y., Ohkuma, M., Suzuki, K. & Haruta, S., 2013. Candidatus Methanogranum caenicola: A novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes. Environ. 28(2), 244-250. Doi: 10.1264/jsme2.me12189. [ Links ]
Johnson, K.A. & Johnson, D.E., 1995. Methane emissions from cattle. J. Anim. Sci. 73(8), 2483-2492. Doi: 10.2527/1995.7382483X. [ Links ]
Kittelmann, S., Seedorf, H., Walters, W.A., Clemente, J.C., Knight, R., Gordon, J.I. & Janssen, P.H., 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal, and eukaryotic microorganisms in rumen microbial communities. PloS One. 8(2), e47879-e47889. Doi: 10.1371/journal.pone.0047879. [ Links ]
Knight, T., Ronimus, R.S., Tootill, D.D.C., Naylor, G., Evans, P., Molano, G., Smith, A., Tavendale, M., Pinares-Patino, C.S. & Clark, H., 2011. Chloroform decreases rumen methanogenesis and methanogen populations without altering rumen function in cattle. Anim. Feed Sci. Tech. 166-167, 101-112. Doi: 10.1016/j.anifeedsci.2011.04.059 [ Links ]
Kumar, S., Indugu, N., Vecchiarelli, B. & Pitta, D.W., 2015. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front. Microbiol. 6, 781-789. Doi: 10.3389/fmicb.2015.00781. [ Links ]
Leahy, S.C., Kelly, W.J., Altermann, E., Ronimus, R.S., Yeoman, C.J., Pacheco, D.M., Li, D., Kong, Z., McTavish, S., Sang, C., Lambie, S.C., Janssen, P.H., Dey, D. & Attwood, G.T., 2010. The genome sequence of the rumen methanogen, Methanobrevibacter ruminantium, reveals new possibilities for controlling ruminant methane emissions. PLoS One. 5(1), e8926-e8942. Doi: 10.1371/journal.pone.0008926. [ Links ]
Lima, P.R., Apdini, T., Freire, A.S., Santana, A.S., Moura, L.M.L., Nascimento, J.C.S., Rodrigues, R.T.S., Dijkstra, J., Garcez Neto, A.F., Queiroz, M.A.A. & Menezes, D.R., 2019. Dietary supplementation with tannin and soybean oil on intake, digestibility, feeding behavior, ruminal protozoa, and methane emission in sheep. Anim. Feed Sci. Tech. 249, 10-17. Doi: 10.1016/j.anifeedsci.2019.01.017. [ Links ]
Liu, J., Zhang, M., Xue, C., Zhu, W. & Mao, Y., 2016. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J. Dairy Sci. 99, 9668-9681. Doi: 10.3168/jds.2016-11398. [ Links ]
Mccaughey, W.P., Wittenberg, K. & Corrigan, D., 1999. Impact of pasture type on methane production by lactating beef cows. Can. J. Anim. Sci. 79(2), 221-226. Doi: 10.4141/a98-107. [ Links ]
Moss, A.R., Jouany, J.P. & Newbold, J., 2000. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 49(3), 231-253. Doi: 10.1051/animres:2000119. [ Links ]
Noel, S.J., H0jberg, O., Urich, T. & Poulsen, M., 2016. Draft genome sequence of 'Candidatus Methanomethylophilus' sp. 1R26, enriched from bovine rumen, a methanogenic archaeon belonging to the Methanomassiliicoccales order. GenomeA. 4(1), e01734-e01735. Doi: 10.1128/genomeA.01734-15. [ Links ]
Pedreira, M.D.S., Oliveira, S.G.D., Primaves, O., Lima, M.A.D., Frighetto, R.T.S. & Berchielli, T.T., 2013. Methane emissions and estimates of ruminal fermentation parameters in beef cattle fed different dietary concentrate levels. Rev. Bras. Zootecn. 42(8), 592-598. Doi: 10.1590/S1516-35982013000800009. [ Links ]
Peng, X., Wilken, S.E., Lankiewicz, T.S., Gilmore, S.P., Brown, J.L., Henske, J.K., Swift, C.L., Salamov, A., Barry, K., Grigoriev, I.V., Theodorou, M.K., Valentine, D.L. & O'Malley, M.A., 2021. Genomic and functional analyzes of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat. Microbiol. 6(4), 499-511. Doi: 10.1038/s41564-020-00861-0. [ Links ]
Philippeau, C., Lettat, A., Martin, C., Silberberg, M., Morgavi, D.P., Frelay, A., Berger, C. & Nozière, P., 2017. Effects of bacterial direct-fed microbials on ruminal characteristics, methane emission, and milk fatty acid composition in cows fed high- or low-starch diets. J. Dairy Sci. 100(4), 2637-2650. Doi: 10.3168/jds.2016-11663. [ Links ]
Seedorf, H., Kittelmann, S. & Janssen, P.H., 2015. Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Appl. Environ. Microbiol. 81(3), 986-995. Doi :10.1128/aem.03018-14. [ Links ]
Shi, W.B., Moon, C.D., Leahy, S.C., Kang, D.W., Froula, J., Kittelmann, S., Fan, C., Deutsch, S., Gagic, D., Seedorf, H., Kelly, W.J., Atua, R., Sang, C., Soni, P., Li, D., Pinares-Patino, C.S., McEwan, J.C., Janssen, P.H., Chen, F., Visel, A., Wang, Z., Attwood, G.T. & Rubin, E.M., 2014. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24(9), 1517-1525. Doi: 10.1101/gr.168245.113. [ Links ]
Sirohi, S.K., Chaudhary, P.P., Singh, N., Singh, D. & Puniya, A.K., 2013. The 16S rRNA and mcrA gene based comparative diversity of methanogens in cattle fed on high fibre-based diet. Gene. 523(2), 161-166. Doi: 10.1016/j.gene.2013.04.002. [ Links ]
Tajima, K., Nagamine, T., Matsui, H., Nakamura, M. & Aminov, R.I., 2001. Phylogenetic analysis of archaeal 16S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol. Lett. 200(1), 67-72. Doi: 10.1016/s0378-1097(01)00201-4. [ Links ]
Tomkins, N.W., Denman, S.E., Pilajun, R., Wanapat, M., McSweeney, C.S. & Elliott, R., 2015. Manipulating rumen fermentation and methanogenesis using an essential oil and monensin in beef cattle fed a tropical grass hay. Anim. Feed Sci. Tech. 200, 25-34. Doi: 10.1016/j.anifeedsci.2014.11.013. [ Links ]
Wallace, R.J., Rooke, J.A., Mckain, N., Duthie, C.A., Hyslop, J.J., Ross, D.W., Waterhouse, A., Watson, M. & Roehe, R., 2015. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics. 16(1), 1-14. Doi: 10.1186/s12864-015-2032-0. [ Links ]
Wang, C., Zhang, C., Yan, T., Chang, S., Zhu, W., Wanapat, M. & Hou, F., 2019. Increasing roughage quality by using alfalfa hay as a substitute for concentrate mitigates CH4 emissions and urinary N and ammonia excretion from dry ewes. J. Anim. Physiol. An. N. 00, 1-10. DOI: 10.1111/jpn.13223. [ Links ]
Wang, Z., Elekwachi, C.O., Jiao, J.Z., Wang, M., Tang, S.X., Zhou, C.S., Tan, Z.L. & Forster, R.J., 2017. Investigation and manipulation of metabolically active methanogen community composition during rumen development in black goats. Sci. Rep-UK. 7(1), 422-435. Doi: 10.1038/s41598-017-00500-5. [ Links ]
Yang, C.J., Mao, S.Y., Long, L.M. & Zhu, W.Y., 2012. Effect of disodium fumarate on microbial abundance, ruminal fermentation, and methane emission in goats under different forage:concentrate ratios. Animal. 6(11), 1788-1794. Doi: 10.1017/S1751731112000857. [ Links ]
Zhu, Z., Noel, S.J., Difford, G.F., Al-Soud, W.A., Brejnrod, A., S0rensen, S.J., Lassen, J., Lovendahl, P. & Hojberg, O., 2017. Community structure of the metabolically active rumen bacterial and archaeal communities of dairy cows over the transition period. PloS One. 12(11), e0187858-e0187881. Doi: 10.1371/journal.pone.0187858. [ Links ]
Submitted 13 February 2023
Accepted 12 April 2023
Published 12 July 2023
# Corresponding author: narenhualaoshi@163.com














