Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.52 n.6 Pretoria 2022
http://dx.doi.org/10.4314/sajas.v52i6.02
MC4R gene polymorphisms for classification of growth efficiency and carcass measurements in two rabbit breeds in Egypt
Hend A. RadwanI, #; Ahmed I. AteyaI; Eman A. Abo ElfadlI; Shimaa A. SakrI; Mohamed M. FoudaI; Ragab A. DarwishI; Adel E. El-DesokyII
IDepartment of Husbandry and Development of Animal Health, Faculty of Veterinary Medicine, Mansoura University, Mansoura 35516, Egypt
IIDepartment of Waterfowl and Rabbit Research, Animal Production Research Institute, Dokki, Giza, Egypt
ABSTRACT
Melanocortin 4 receptor (MC4R), a protein derived from the MC4R gene, is involved in feed intake, metabolism control, and body weight regulation in humans. The purpose of this research was to explore MC4R polymorphisms alongside metabolic marker changes and their relationship with growth and carcass measurements in rabbits. Using synthetic line V (V-line) and Baladi Black rabbit breeds (60 rabbits per breed), blood samples were collected for DNA extraction and biochemical analysis. The polymerase chain reaction (PCR) product of MC4R (493 bp) revealed five nonsynonymous single nucleotide polymorphisms (SNPs; submitted to GenBank with accession numbers gb|MT832144|, gb|MT832145|, and gb|MT832146|). Four SNPs were characteristic of the V-line breed, and one was characteristic of the Baladi Black. For classification of the defined SNP-dependent groups within and between breeds, a discriminant analysis model correctly classified a percentage of cases with the following predictor variables: 90.8% for body weight at 5-14 weeks of age; 85% for feed consumption, daily feed intake, and feed conversion ratio; and 93.3% for carcass measurements (for which hind part weight, liver weight, and liver percentage were the best predictors in both breeds). There were significant differences between and within V-line and Baladi Black breeds in agreement with metabolic biochemical marker profiles and the defined SNPs. The identified SNPs in the MC4R gene and profile of the investigated metabolic biomarkers could be used as candidates and reference for the effective characterization of the two rabbit breeds. This study could therefore facilitate the introduction of marker-assisted selection for growth performance characteristics in rabbits.
Keywords: discriminant analysis, feed conversion efficiency, Melanocortin 4 receptor, slaughter traits
Introduction
Wild rabbits are commonly considered to have been first domesticated in 600 A.D by French medieval monks (Doherty & Driscoll, 2017). The domestic rabbit (Oryctolagus cuniculus domesticus) is a subspecies of the European rabbit (Oryctolagus cuniculus), which belongs to the Leporidae family, of the order Lagomorpha. The Oryctolagus genus includes the European rabbit species as well as its descendants, the world's 305 domestic rabbit breeds. European rabbits can be found in the wild on all continents, with the exception of Asia and Antarctica (DAD-IS, 2017).
Recently, domestic rabbits have been proposed as a good substitute source of dietary protein for the rising human populations in developed countries (Dalle Zotte, 2014; Ezema & Eze, 2015; Trocino, 2019). According to the Food and Agriculture Organization of the United Nations (FAO) China, North Korea, Spain, Egypt, and Italy were the five leading rabbit meat producers globally in 2017 (FAO, 2019). Rabbit meat is healthy and nutritious; it is commonly given to children and the elderly because of its nutritional benefits and efficient digestibility (Dalle Zotte & Szendro, 2011; Dalle Zotte, 2014). Rabbit meat is characterized by a high protein content (~22%), a large proportion of essential amino acids, a loin lipid content of ~1.8g/100g meat (Dalle Zotte & Szendro, 2011), and high vitamin B content and it is low in fat, cholesterol, and sodium (Ramirez et al., 2006; Ezema & Eze, 2015). Moreover, rabbit meat qualifies as one of the most valuable meat sources due to its effectiveness in dietary manipulation, as well as a promising improvement in oxidative stability and "functional" properties (Pla et al., 2004; Dalle Zotte & Szendro, 2011; Dalle Zotte et al., 2016; Martins et al., 2018). Indeed, as a tender and nearly cholesterol-free white meat, rabbit meat could potentially replace chicken meat (Dalle Zotte, 2014). Overall, the rabbit has high value as a meat product.
In 1981, the V-line (or line V) was developed as a synthetic line in Spain (Polytechnic University of Valencia) by crossing the progeny of four specific maternal lines chosen to improve litter size at weaning (Estany et al., 1989). A maternal synthetic line is created by improving doe-related traits such as litter weights and sizes, as well as milk yield, which are employed as selection criteria in the development of maternal rabbit lines (Estany et al., 1989; Gómez et al., 1996; Rochambeau et al., 1998; Baselga, 2004).
A variety of characteristics related to climate adaptation and litter size at weaning dictated the decision to select line V in this study. Valencia seems to have a long selection history (García & Baselga, 2002), and its climate is akin to that of Egypt's Nile delta. The V line breed has also been examined in hot climates such as Adana in Turkey and Zagazig in Egypt, where it surpassed other exotic breeds (Yamani, 1994) according to the first international hot climate rabbit development conference in Cairo (Yamani,1994) and the sixth world rabbit congress in Toulouse (Testik, 1996). New reports in Saudi Arabia for the V-line have reaffirmed the line's superior heat stress tolerance (Khalil et al., 2002). This line is also notable for having a higher litter size at weaning.
According to the Egyptian Ministry of Agriculture's Poultry Breeding Section, the Baladi rabbit breed originated from Egypt as a consequence of crossbreeding between native rabbits and the exotic Flemish Giant breed for many generations at research stations to develop a heat-resistant, meat-producing rabbit that could endure Egypt's harsh climate (Badawy, 1975; Galal & Khalil, 1994). This selection approach for the adapted lines for the Egyptian climate resulted in the establishment of three native Baladi strains: Baladi Red, Baladi White, and Baladi Black (Khalil, 2011).
Growth performance and carcass traits are of high importance in the overall breeding objective of rabbit selection programs. However, the effects of a large number of known gene polymorphisms associated with these traits require assessment to improve breeding operations (Migdal et al., 2019). SNP genetic markers have revolutionized previous achievements in conservation decisions, biodiversity assessment, and genetic characterization of breeds (Groeneveld et al., 2010). Few studies using genotypes dependent on single nucleotide polymorphisms (SNPs) of candidate genes have been undertaken to enhance the genetic potential of rabbits for human food purposes (El-Sabrout, 2017). The diversity of rabbit breeds coupled with tailored breeding strategies could potentially enhance the efficiency of commercial meat production (Piles et al., 2004).
Molecular genetic markers (e.g., restriction fragment length polymorphisms, RFLP; SNP) can be used in rabbit management to improve selection and mating systems by favouring the preferable genotype; and thereby reducing kindling intervals among generations (Hirose et al., 2014). Using these markers could also improve characteristics such as reproductive performance, growth and development, quality of meat, and milk production (Dekkers, 2004). Molecular markers may also be used to increase the precision of selection and, therefore, the genetic progress of major economic traits. Several previous investigations have demonstrated correlations between candidate gene polymorphisms and meat characteristics in rabbits. Many polymorphisms have been correlated with rabbit body weight (BW), e.g., myostatin SNPs (Sternstein et al., 2014), growth hormone (GH) (Fontanesi et al., 2012a), GH receptor (Zhang et al., 2012), and insulin-like growth factor 2 (IGF-2) (Fontanesi et al., 2012b). Moreover, Zhang et al. (2013) reported that SNPs within the fat mass and obesity-associated (FTO) gene are linked to the longissimus lumborum muscle having higher intramuscular fat content.
Melanocortin 4 receptor (MC4R), a member of the G-protein-coupled receptor family, is expressed in the human hypothalamus. It is known to be involved in food intake as well as the regulation of metabolism and body weight (Li & Li, 2006). Polymorphisms in the MC4R gene have been associated with growth performance in domestic animals such as pigs (Houston et al., 2004; Meidtner et al., 2006), cattle (Zhang et al., 2009), sheep (Song et al., 2012), and chickens (Zhou et al., 2012). It was established that the MC4R gene could be a potential candidate for production traits; however, the possible association of MC4R gene polymorphisms in rabbits has rarely been reported. This study therefore aims to explore the association between MC4R gene polymorphisms and growth traits, carcass traits, feed intake, and feed conversion ratio in V-line and Baladi Black rabbit breeds using a PCR-DNA sequencing approach, to subsequently identify whether there are associations between the SNPs identified in the MC4R region and serum levels of various metabolic biochemical markers including growth hormone (GH), leptin (Lep), and thyroid stimulating hormone (TSH) in these breeds.
Materials and Methods
This study was conducted from November, 2017 to March, 2019 on rabbits raised at the El-Serw Experimental Station, which belongs to the Animal Production Research Institute (Agricultural Research Center, Ministry of Agriculture, Egypt) from birth to weaning at 5 weeks of age. Thereafter, growth and feed performance records were collected from 120, five-week-old rabbits from the V-line (n = 60) and Baladi Black (n = 60) breeds until 14 weeks of age whereafter the animals were slaughtered and carcass traits were recorded. The collection of samples and care of rabbits used in this study followed the guidelines of Mansoura University and the protocol of the study was approved by the Research Ethics Committee, Faculty of Veterinary Medicine, Mansoura University (code R/21).
The rabbits were raised in an open-sided hutch. Breeding animals were housed separately in galvanized wire cages (40 χ 60 χ 50 cm3) fitted with a nipple drinking system and a manual feeder.
Diazinon (20%) was routinely used to wash and purify the hutches before each kindling. Cages and nest boxes were regularly cleansed to get rid of mange. Affected animals were handled with injectable Ivomac (Ivermectin injection 2%) biweekly for non-pregnant does and young, and Benzylbenzoate cream for pregnant does. Urine and faecal pellets on the floor of the cages were cleaned each morning.
The environmental temperature was preserved as far as possible between 16 °C and 24 °C. Efficient ventilation and fresh air were provided by fans to minimize the accumulation of ammonia in the building. Photoperiod was set at 16 light vs 8 dark in the rabbitry.
A tubular-shaped, pelletized (4 mm diameter, 9 mm length) commercial ration was utilized. The ration formulated according to Zaghloul et al. (2019), contained 24% soybean meal, 23% barley, 21% berseem hay, 19% wheat bran, 18.01% crude protein 13% yellow corn, 11.5% crude fiber, 2.5% fat, 1% limestone, 0.5% table salt, 14 kg di-calcium phosphate/ton, 1 kg anti-toxicity/ton, 1 kg anti-coccidial/ton, and 1 kg of a mineral mixture/ton, as per the National Research Council (NRC,1977).
The rabbits were supplied with safe, fresh water at all times. Prophylactic antibiotics and anti-coccidial medications were applied to the water for 3-5 days until weaning. Kits were injected every two weeks with multi-vitamins to mask any shortages and operate against declining immunity arising from weaning (Lebas, 2000). In addition, injectable vitamin E and selenium were also used for fertility enhancement. Kits were vaccinated at three and six weeks of age, respectively, against pasteurellosis and infectious rabbit haemorrhagic disease as well as a booster dose at ten weeks of age, according to the history of the El-Serw Animal Production Research Station vaccination program and area circumstances.
Mating was done at random, with the exception of parent-offspring, full-sibling, and half-sibling mating. Each doe was mated with the appropriate buck of the same breed in a ratio of 1 buck to 3 does from each breed. Ten days after mating, each doe was palpated to confirm pregnancy, and those found to be non-pregnant were reintroduced to the same buck. Attached to the doe's cage was a metal nest box (40 χ 40 χ 40 cm3) for kindling and nursing kits. The nest boxes were supplied with a thick layer of rice straw on the 27th day of pregnancy, which was placed in the bottom of the nest box to assist the doe in creating a warm and comfortable nest for her offspring. At weaning (five weeks), litters were ear-tagged, and separated into cages, each cage containing five litters to start the fattening period until marketing (14 weeks).
During the fattening period, male and female rabbits were weighed weekly to determine their body weight (BW). Weight gain (WG) was expressed as the difference between the current and previous weight relative to the subsequent period of time. Average daily gain (ADG) was defined as the weight gain over the computed number of days.
Rabbits were supplied with a precisely weighed amount of feed per day to calculate feed intake (FI). At the end of the day, the residual feed was excluded and the intake amount was determined by the difference according to the method of Wagner et al. (1983). The feed conversion ratio (FCR) was determined by calculating FI per kg and BW gain per kg according to the method of Iyayi & Odueso (2003).
Rabbits were fasted for 12 h at the end of the fattening period and weighed before being slaughtered. They were subsequently euthanized via IP injection of a substantial dose (20 to 60 mg/kg) of sodium thiopental for organ and body part collection (Archimedes, France) according to Hellebrekers et al. (1990). The skin of the slaughtered animals was removed and dressed, and the hot carcasses were separately weighed and examined, along with the head and giblets.
The dressing percentage (dressing %) was calculated as follows:
dressing percentage = 100 χ hot carcass (giblets weight + head weight + carcass weight) / live BW, where giblets weight = kidney weight + heart weight + liver weight.
The ratio of body organs (e.g., kidneys, heart, abdominal fat, mid parts, foreparts, hind parts, liver, and head) to carcass weight was estimated according to the method of Szendrõ et al. (2010).
Blood samples were collected aseptically before slaughter from 120 rabbits (60 of each breed). After washing and disinfection, approximately 2 ml of blood was obtained from a rabbit's ear vein using a piece of cotton filled with xylol in tubes containing EDTA for DNA extraction. Tubes without anticoagulants were used to obtain serum by centrifugation. The serum samples were subsequently placed in a cold gel icebox, which was sealed and stored at 4 °C prior to a hormonal assay.
Genomic DNA was extracted from whole blood using a Gene JET Whole Blood Genomic DNA Extraction Kit following the manufacturer's instructions (Thermo Scientific, Lithuania). The quality, purity, and concentration of DNA were assessed using a Nanodrop before further analysis.
Polymerase chain reaction (PCR) was used to amplify fragments of the MC4R gene, which spanned exons I and II, with an expected amplicon size of 493 bp (Fontanesi et al., 2013). The primers used were: forward: 5'-CCATTGCAGTGGACAGGTATT-3'; reverse: 5'-TCCGGAGTGCATAAATGAGA-3'. The PCR mixture (50 μ!) contained DNA (3 μ!), H2O (double-distilled water, 21 μ!), PCR master mix (25 μ!; Jena Bioscience, Germany), and forward and reverse primers (0.5 μl of each). The final reaction mixture was placed in a thermal cycler, and the PCR program was conducted with an initial denaturation at 95 °C for 3 min, followed by 30 cycles of 95 °C for 1 min for DNA denaturation, annealing at 55 °C for 1 min, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. The samples were then maintained at 4 °C; representative PCR results were confirmed by agarose gel electrophoresis, and then fragment patterns were visualized under ultraviolet light using a gel documentation system.
PCR products with target bands were used for DNA sequencing. These products were sequenced in forward and reverse directions with an ABI 3730XL DNA sequencer (Applied Biosystems, USA) according to the enzymatic chain terminator technique developed by Sanger et al. (1977). DNA sequencing data were analysed using Chromas 1.45 and BLAST 2.0 (Altschul et al., 1990). Differences were classified as SNPs on the basis of a comparison between the PCR products of the MC4R gene and the reference sequence available in GenBank. On the basis of an alignment of sequences, variation in the amino acid sequences of MC4R genes among the breeds was identified using MEGA4 (Tamura et al., 2007).
Using an ELISA analyser, growth hormone (GH) was assessed by adding 100 μΙ of weak dilution antibody to each well of a plate, which was then stored at room temperature (20-25 °C) for 1 h, after which 200 μ! of blocking solution was added for 30 min, and the plate was washed multiple times. Subsequently, 100 steps were applied to each well, which was then maintained for 1 h at room temperature. Another 100 μΙ of dilution antibody was then added to the wells for 1 h at room temperature. The wells were washed, containing 100 μΙ of the well's TMP substrate solvent, and were positioned in a dark place at room temperature for 15 min, including 100 μl of the well suspension solution. Color absorption was then calculated on a plate reader at approximately 450 nm (Medgyesi et al., 1975).
Leptin hormone (Lep) was evaluated using IMMULITE and an IMMULITE 1000 Analyzer for the quantitative estimation of leptin in serum, according to the method of Tian (2005). A solid-stage, chemical (enzyme)-labelled chemiluminescent competitive immunoassay was used, with the bead (solid stage) coated in anti-leptin polyclonal rabbit antibody. The fluid phase was composed of leptin-conjugated alkaline phosphatase (bovine calf intestine).
Thyroid stimulating hormone (TSH) concentration was determined according to the method of Baloch et al. (2003). Kits from Siemens Health Diagnostic (USA) were used with Speedy TSH IMMULITE/IMMULITE 2000, a powerful immunochemical assay.
Data were arranged, compiled, and analysed using the statistical package, SPSS version 23 (USA). The sample size was calculated using the following equation based on the population size:
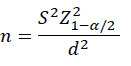
where: S2 = population variance; 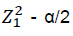 = critical value for significance level which equals 1.96 for 95% confidence level; d = level of error.
= critical value for significance level which equals 1.96 for 95% confidence level; d = level of error.
A chi-squared test was used to evaluate the frequencies of MC4R SNPs across various classes of the V-line (GV1, GV2, GV3, and GV4) and Baladi Black (GB1 and GB2) breeds. A linear discriminant analysis (LDA) (SPSS, 2015) was then used to evaluate the significance of various determinants and classify gene group SNPs as the dependent variables, with BW, WG, FI, FCR, and carcass traits as the independent attributes. The discriminating statistical model was:
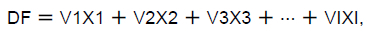
where DF= the discriminating grouping function (score) for grouping variables; V = the standardized discriminant or loading coefficient for predictors; X = the respondent's score for the predictors; and I is the number of predictor variables.
The coefficients of discriminant equation, V, or the idealized type, beta, represent the partial contribution of each predictor to the mechanism of discrimination. Finally, a one-way ANOVA was used to compare the means of BW, WG, FI, FCR, and carcass traits among MC4R groups, dependent on SNP classes. All data were represented as means ± SE; P <0.05 was considered statistically significant.
Results and Discussion
In this study, DNA sequencing of the MC4R gene (493 bp) revealed five SNPs (submitted to GenBank with accession numbers gb|MT832144|, gb|MT832145|, and gb|MT832146|; Table 1). Four non-synonymous SNPs were characteristic for the V-line breed; rabbits that harboured the A47C SNP, the A50C SNP, the A43G SNP, the A44T SNP, and those that did not harbour the identified SNPs, are hereafter respectively referred to as GV1MC4R, GV2MC4R, GV3MC4R, and GV4MC4R. For Baladi Black rabbits, one identified synonymous SNP (G333A) was identified and this group is hereafter referred to as GB1MC4R; whereas Baladi Black rabbits that did not exhibit the identified SNP are hereafter referred to as GB2MC4R.
Chi-square tests revealed a difference in the frequency of MC4R gene SNPs between the two breeds (Table 1). Nucleotide sequence variation for the MC4R gene between the two breeds, as well as between the breeds and reference sequences available in GenBank (HF970577.1), confirmed all five identified SNPs. The identification of some SNPs specific to each breed is probably related to the founder effect associated with origin, history, evolution, and genetic constituents of each breed (Carneiro et al., 2011; Ateya et al., 2021).
The MC4R gene is known to play an active role in food intake, metabolic control, and body weight in humans (Li & Li, 2006). MC4R is a descendant of the G-protein paired receptor family, which plays a key role in managing rodent feeding behaviour (Wardlaw, 2001). Indeed, variations of MC4R were found to be consistent with genetic human obesity (Farooqi & O'Rahilly, 2006). Previous studies have focused on the association between the MC4R gene and control of eating behavior in humans, mice, and pigs (El-Sabrout, 2017). Furthermore, MC4R gene mutations in many animal species are correlated with carcass quality and growth, e.g., in chickens (Wang et al., 2009) and cattle (Zhang et al., 2009). According to our results, the identified SNPs in the MC4R gene were associated with growth traits in the two breeds enabling for marker assisted selection within and between breeds.
The association between MC4R gene polymorphisms and productive traits in rabbits has not previously been studied in detail. However, El-Sabrout & Soliman (2017) reported an association between SNPs of the MC4R gene and FI in V-line rabbits and El-Sabrout & Aggag (2018) reported an association between MC4R gene polymorphisms and carcass quality in rabbits. According to the results, the authors reported that identified SNPs were associated with feed intake. Fontanesi et al. (2013) also found that a missense mutation in the rabbit MC4R gene was associated with finishing weight in a meat rabbit line. Additionally, Osaiyuwu et al. (2020) reported that a MC4R gene polymorphism was associated with body weight in some breeds of rabbit. Previous studies have investigated the association of MC4R gene polymorphisms and their association with growth performance in rabbit breeds using other genetic markers (e.g., RFLP and single-strand conformational polymorphism, SSCP) (Jiang et al., 2008; Nahácky et al., 2018).
Due to their abundance and distribution across the genome, high automation, and multiplex ability, SNP markers have revolutionized previous achievements in conservation decisions, biodiversity assessment, and genetic characterization of breeds. Although these markers are applied intensively across other production animal species, the same has not been true for rabbits. Studies like this one might assist in improving SNP-based applications for rabbits (Groeneveld et al., 2010). Jiang et al. (2008), for example, reported SNP markers in the coding sequence of the MC4R gene detected by the PCR-SSCP and DNA sequencing methods. By means of a general linear model analysis for the effect of genotypes on performance traits, these authors demonstrated that an A/G genotype was associated with body weight, eviscerated weight, and feed conversion efficiency (P <0.05) but not associated with cooking loss (P >0.05). Nahácky et al. (2018) also reported a MC4R gene polymorphism that was associated with production traits in rabbit.
The hormonal assay revealed that GH and TSH varied greatly among the V-line and Baladi Black breeds with various MC4R SNPs. GB2MCR4 showed the lowest GH levels (1.26 ng/ml) and the highest TSH levels (1.21 μΙ/ml). The relationship between the SNP genotypes and the corresponding metabolic profiles holds potential for explaining the phenotypic variation in the growth performance between the two breeds. On the other hand, there was no substantial difference in leptin hormone among breeds (Table 2). A significant effect of breed on T3 and T4 has been recorded in livestock; for instance, Ghanem et al. (2016) concluded that the choice of broiler breed may have a significant impact on serum T3 and T4 levels, and these findings may be reflected in growth and body composition. Leptin hormone levels in Baladi Black rabbits were found to be insignificantly higher than those in NZW rabbits for both does and bucks (El-Werdany et al., 2016).
Studied traits were entered into a DF model using a stepwise method. All variables except DFI, WG, and DWG were selected for use in the equation as predictors for the allocated groups based on identified SNPs in the MC4R gene. For BW at 5-14 weeks of age, there was a genetic variation between the V-line and Black Baladi breeds, with GV1MC4R, GV2MC4R, GV3MC4R, and GV4MC4R SNP-allocated groups showing a higher BW compared to the Baladi Black (GB1MC4R, GB2MC4R) breed. In the same respect, there was a genetic variation within the V-line and Black Baladi breeds in BW, where GV1MC4R and GV2MC4R SNP-allocated groups had a higher BW than the others. The GB1MC4R group showed a higher BW than GB2MC4R in the Black Baladi breed (Table 3). The observed results were consistent with those of El-Sabrout & Soliman (2018), where authors analysed the relationship between MC4R gene polymorphisms and rabbit feed intake. Results revealed that identified SNPs were correlated with feed intake, which facilitate marker assisted selection (MAS). Their findings can also be used to select rabbits for high body weight at a marketing age. Additionally, it was established that MC4R gene polymorphisms were associated with finishing weight (Fontanesi et al., 2013). MCR4 was associated through sequencing with high BW at market for Alexandria and V-line breeds by El-Sabrout & Aggag (2017). Conversely, Shan et al. (2020) found no significant association between the 19 detected SNPs of the MC4R gene and body weight traits of Hu sheep; however, they noticed two SNPs (g.706 CA and g.732 CG) that were significantly associated with adult female body height traits.
Weekly body weight during the fattening period (5 to14 weeks of age) was used as a predictor in the LDA for classification of the SNP groups of MC4R. Correctly classified cases are the number of cases that have been correctly classified to the appropriate category in terms of the determinant (BW, 5-14 weeks). Overall, 90.8% of cases were correctly classified by the model, with 100% (17/17) for GV1MC4R, 78.26% (18/23) for GV2MC4R, 84.62% (11/13) for GV3MC4R, 71.43% (5/7) for GV4MC4R, 97.96% (48/49) for GB1MC4R, and 90.09% (10/11) for GB2MC4R. It was also shown that BW10 was the best predictor for function 1, BW6 was the best predictor for function 2, BW8 was the best predictor for function 3, BW13 was the best predictor for function 4, and BW11 was the best predictor for function 5 (estimates of 0.369, 0.904, 0.719, -1.960, and -1.313, respectively) (Table 4). On the basis of these results, rabbits with weight problems can be eliminated early in the fattening period, e.g. up to the sixth week after birth, as in the second formula.
The SNP-dependent allocated groups of the two studied breeds were strongly associated with carcass traits; there were genetic differences between and within SNP-allocated groups from both the V-line and Baladi Black rabbit breeds (Table 5). The variation/diversity in these genetic resources may be beneficial for future breed management and improvement.
All carcass traits except forepart weight, foreparts %, and middle parts % were used as predictors in LDA for the classification of SNP groups. In terms of determinant (carcass traits), correctly classified cases corresponded to the number of cases that had been correctly classified into the correct category. Overall, the model correctly classified 93.3% of cases, with 100% (17/17) for GV1MC4R, 91.3% (21/23) for GV2MC4R, 69.23% (9/13) for GV3MC4R, 85.71 % (6/7) for GV4 MC4R, 100% (49/49) for GB1 MC4R, and 81.82% (9/11) for GB2MC4R. It was also shown that liver weight was the best predictor for function 1, hind part weight was the best predictor for function 2, liver % was the best predictor for function 3, and hind part weight was the best predictor for functions 4 and 5 (estimates of -3.315, -1.559, 7.033, 8.130, and -11.260, respectively) (Table 6). These results demonstrate the efficiency of determining the breeding method and mating system within rabbit farms by selecting rabbits with desirable traits to be the parents of future generations. Xuemei et al. (2006) discovered an association between various genotypes of the chicken MC4R gene and body weight, growth, and carcass traits. They revealed that the body weight, carcass weight, and leg muscle weight of the BB were dramatically higher than those of the AA and AB genotypes (P <0.05), but the difference among the three genotypes did not attain the significance level (P >0.05). The afore-mentioned authors also concluded that the MC4R gene is the most likely candidate gene for chicken growth and carcass traits.
In terms of feed intake, DFI, and FCR there was substantial variation between the V-line and Black Baladi class SNPs, with the Black Baladi SNPs outperforming the V-line SNPs at most intervals for these traits (Table 7). Jiang et al. (2008) reported that allele A of the MC4R gene was the predominant allele in five studied rabbit breeds; therefore, the AA genotype level was greater than the AG genotype. GLM analysis for the influence of genotypes on productive traits demonstrated that the genotype of AG was truly associated with BW, eviscerated weight, and feed conversion efficiency. G/A 1426 MC4R mutations were genotyped using real-time PCR approach by Piórkowska et al. (2010) in 1191 gilts of five breeds to determine the impact of missense mutations on meat quality, carcass composition, and growth traits. They noticed that the A allele was linked to higher daily feed intake (AA - 2.51 kg; GG - 2.31 kg in the Putawska breed, P <0.05), daily gain, and backfat thickness (AA -1.67 cm, GG -1.52 cm in PL, P <0.01). Reduced lean meat content was exhibited by the AA genotype.
The effect of MC4R on rabbit FI indicates that it may be a significant genetic marker for the growth-related features of rabbits. Moreover, nucleotide sequence assessment of whole amplified fragments and evaluation of the standard errors and means of individual rabbits' BW at 9 weeks of age have confirmed that significant correlations exist between MC4R SNPs and high rabbit BW at marketing age (El-Sabrout & Aggag, 2017; El-Sabrout, 2017). Interestingly, our study elicited novel SNPs in the MC4R gene associated with growth traits in rabbits. El-Sabrout & Soliman (2018) explained the relationship between two parts of the melanocortin gene (MC4R-1 and MC4R-2) and FI for V-line rabbits (low and high FI); they observed that the alignment of sequence data from each group revealed that there is a variation detected in MC4R-1 at nucleotide 35 (T-G) (sense mutation) and another variation was detected in MC4R-2 gene at nucleotide 19 (T-C) (sense mutation) for high feed intake rabbits. The MC4R gene's function is substantially altered as a result of these sense mutations, which modify certain amino acids. The average daily feed intake findings showed that the high feed intake group had a substantially higher feed intake than the V-line rabbits' high feed intake group. The indicated mutations, as well as the assessment of daily feed intake means, demonstrated a substantial link between the MC4R polymorphism and rabbit feed intake. The same association with BW, obesity, and development was reported by Nahácky et al. (2018). On the other hand, Dvoáková et al. (2011), found no effect of the p.Asp298Asn mutation of the MC4R gene on feed intake, feed conversion, and growth rate in a Czech pig population.
Feed intake, DFI, and FCR were entered in the DF model using a stepwise method; FI and FCR were selected for use in the equation as predictors. FCR and FI were also used as predictors in LDA for classification of the SNP groups of MRC4. Correctly classified cases are the number of cases that have been correctly classified to the appropriate category in terms of the determinants (FCR & FI). Overall, the model correctly classified 85% of cases, with 64.71% (11/17) for GV1MC4R, 95.65% (22/23) for GV2MC4R, 92.31% (12/13) for GV3MC4R, 5.6% (2/7) for GV4MC4R, 89.79% (44/49) for GB1MC4R, and 90.91% (10/11) for GB2MC4R. It was also shown that breed was the best predictor for function 1; FI 5-14 was the best predictor for functions 2, 3, 4, and 5 for both FI and FCR; followed by FI 8-11 for functions 2, 3, and 5; and then FI 5-8 for function 4; followed by FCR 8-11 for functions 2 and 5; and then FCR 5-8 for functions 3 and 4 (Table 8).
MC4R SNP-allocated groups in both breeds had an effect on weight gain and daily weight gain; there was genetic variation between and within the group MC4R SNPs for both V-line and Baladi Black rabbit breeds (Table 9). Animal breeders can use this difference as a preliminary step to improve subsequent generations of rabbits.
Authors' contributions
HAR designed, performed the experiment, and collected data; AIA collected blood samples, performed PCR-DNA Sequencing; EAA analysed the data and interpreted the results; SAS performed the experiment, and collected data; MMF, RAD, and AEE wrote the initial manuscript. The manuscript was revised by all authors, and the final version was accepted before release.
Conflict of interest declaration
The authors declare that they have no conflict of interest.
References
Altschul, S.F., Gish, W., Miller, W., Myers, W., & Lipman, D.J., 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. https://doi.org/10.1016/S0022-2836(05)80360-2 [ Links ]
Ateya, A.I., Hendam, B.M., Radwan, H.A., Abo Elfadl, E.A. & Al-Sharif, M.M., 2021. Using linear discriminant analysis to characterize novel single nucleotide polymorphisms and expression profile changes in genes of three breeds of rabbit (Oryctolagus cuniculus). Comp. Med. 71, 3, 222-234. https://doi.org/10.30802/AALAS-CM-20-000103. [ Links ]
Badawy, A.G., 1975. Rabbit Raising. Central Administration for Agricultural Culture, Ministry of Agriculture, Egypt (2nd Edition, In Arabic), pp. 75. [ Links ]
Baloch, Z., Carayon, P., Conte-Devolx, B., Demers, L.M., Feldt-Rasmussen, U., Henry, J.F., LiVosli, V.A., Niccoli-Sire, P., John, R., Ruf, J., Smyth, P.P., Spencer, C.A., & Stockigt, J.R., 2003. Guidelines Committee, National Academy of Clinical Biochemistry. Laboratory medicine practice guidelines. Laboratory support for the diagnosis and monitoring of thyroid disease. Thyroid: official journal of the American Thyroid Association 13, 1, 3-126. https://doi.org/10.1089/105072503321086962 [ Links ]
Baselga, M., 2004. Genetic improvement of meat rabbits. Programmes and diffusion. In Proc.: 8th World Rabbit Congress, 7-10 September, 2004. Puebla, Mexico, 1-13. [ Links ]
Carneiro, M., Afonso, S., Geraldes, A., Garreau, H., Bolet, G., Boucher, S., Tircazes, A., Queney, G., Nachman, W.M. & Ferrand, N., 2011. The genetic structure of domestic rabbits. Mol. Biol. Evol. 28,1801-1816. https://doi.org/10.1093/molbev/msr003 [ Links ]
DAD-IS [Internet]. 2017. (Domestic Animal Diversity Information System). FAO (Food and Agriculture Organization of the United Nations). [Cited 30 March 2018]. Available at: http://www.fao.org/dad-is/data/en/. [ Links ]
Dalle Zotte, A., & Szendro, Z., 2011. The role of rabbit meat as functional food. Meat Sci. 88,3, 319-331. https://doi.org/10.1016/j.meatsci.2011.02.017 [ Links ]
Dalle Zotte, A., Cullere, M., Alberghini, L., Catellani, P., & Paci, G., 2016. Proximate composition, fatty acid profile, and heme iron and cholesterol content of rabbit meat as affected by sire breed, season, parity order, and gender in an organic production system. Czech J. Anim. Sci. 61, 9, 383-390. https://doi.org/10.17221/24/2016-CJAS [ Links ]
Dalle Zotte, A., 2014. Rabbit farming for meat purposes. Anim. Front. 4, 4, 62-67. https://doi.org/10.2527/af.2014-0035 [ Links ]
Dekkers, J.C., 2004. Commercial application of marker- and gene-assisted selection in livestock: strategies and lessons. J. Anim. Sci. 82, E313-328. https://doi.org/10.2527/2004.8213_supplE313x [ Links ]
Doherty, T.S., & Driscoll, D.A., 2017. Competition in the historical niche: a response to Scheele et al. Trends Ecol. Evol. 33,147-148. https://doi.org/10.1016/j.tree.2017.12.004. [ Links ]
Dvoráková, V., Stupka, R., Sprysl, M., Cítek, J., Okrouhlá, M., Kluzáková, E., & Kratochvílová. H., 2011. Effect of the missense mutation Asp298Asn in MC4R on growth and fatness traits in commercial pig crosses in the Czech Republic. Czech J. Anim. Sci. 56, 4, 176-180.'https://doi.org/10.17221/1305-CJAS [ Links ]
El-Sabrout, K., 2017. Associations between single nucleotide polymorphisms of melanocortin gene and sexual desire behavior in rabbit (Oryctolagus cuniculus). J. Vet. Behav. 19, 69-71. https://doi.org/10.1016/j.jveb.2017.02 [ Links ]
El-Sabrout, K., & Aggag, S.A., 2017. Associations between single nucleotide polymorphisms in multiple candidate genes and body weight in rabbits. Vet. World 10, 136-139. https://doi.org/10.14202/vetworld.2017.136-139 [ Links ]
El-Sabrout, K., & Soliman, F., 2018. Association of single-nucleotide polymorphism of melanocortin gene with feed intake in rabbit (Oryctolagus cuniculus). J. Anim. Physiol. Anim. Nutr. 102, 2, 564-567. https://doi.org/10.1111/jpn.12788 [ Links ]
El-Werdany, I., Saleh, N., Khedr, A., & Hasan, A., 2016. Relationship between lipten hormone concentration and rabbit reproductivity. E.J.R.S. 26, 1, 121-134. https://doi.org/10.21608/ejrs.2016.42040 [ Links ]
Estany, J., Baselga, M., Blasco, A., & Camacho, J., 1989. Mixed model methodology for the estimation of genetic response to selection in litter size of rabbits. Livest. Prod. Sci. 21, 67-76. https://doi.org/10.1016/0301-6226(89)90021-3 [ Links ]
Ezema, C., & Eze, D.C., 2015. Growth performance and cost benefit of weaner rabbits fed diet supplemented with probiotic in the tropics. Pak. J. Nutr. 14, 1, 47-49. https://doi.org/10.3923/pjn.2015.47.49 [ Links ]
Farooqi, I.S., & O'Rahilly, S., 2006. Genetics of obesity in humans. Endocr. Rev. 27, 7, 710-718. https://doi.org/10.1210/er.2006-0040 [ Links ]
Food and Agriculture Organization of the United Nations, (F.A.O., 2019). 2019. FAO database; [accessed 2019 March 5]. http://www.fao.org/3/CA6030EN/CA6030EN.pdf [ Links ]
Fontanesi, L., Dall'Olio, S., Spaccapaniccia, E., Scotti, E., Fornasini, D., Frabetti, A., & Russo, V., 2012a. A single nucleotide polymorphism in the rabbit growth hormone (GH1) gene is associated with market weight in a commercial rabbit population. Livest. Sci. 147, 1-3, 84-88. https://doi.org/10.1016/j.livsci.2012.04.006 [ Links ]
Fontanesi, L., Mazzoni, G., Bovo, S., Frabetti, A., Fornasini, D., Dall'Olio, S. & Russo,V., 2012b. Association between a polymorphism in the IGF2 gene and finishing weight in a commercial rabbit population. Anim. Genet. 43,5, 646652. https://doi.org/10.1111/j.1365-2052.2012.02318.x [ Links ]
Fontanesi, L., Scotti, E., Cisarova, K., Di Battista, P., Dall'Olio, S., Fornasini, D., & Frabetti, A., 2013. A missense mutation in the rabbit melanocortin 4 receptor (MC4R) gene is associated with finishing weight in a meat rabbit line. Anim. Biotechnol. 24, 4, 268-277. https://doi.org/10.1080/10495398.2013.781034 [ Links ]
Galal, E.S.E. & Khalil, M.H., 1994. Development of rabbit industry in Egypt. Cah. Options Méditerr. 8, 43-56. [ Links ]
Garcia, M., & Baselga, M., 2002. Estimation of correlated response on growth traits to selection in litter size of rabbits using a cryopreserved control population and genetic trends. Livest. Prod. Sci. 78, 91-98. https://doi.org/10.1016/S0301-6226(02)00093-3 [ Links ]
Gómez, E.A., Rafel, O., Ramón, J., & Baselga, M., 1996. A genetic study of a line selected on litter size at weaning. In Proc.: 6th World Rabbit Congress, 9-12 July, 1996. Toulouse. France. Vol. 2, 289-292. [ Links ]
Groeneveld, L.F., Lenstra, A.J., Eding, H., Toro, A.M., Scherf, B., Pilling, D., Negrini, R., Finlay, K.E., Jianlin, H., Groeneveld, E., Weigend, S., & Consortium, G., 2010. Genetic diversity in farm animals - a review. Anim. Genet. 41, 6-31. https://doi.org/10.1111/j.1365-2052.2010.02038 [ Links ]
Ghanem, H.M., Ateya, A.I., El Seady, Y.Y., Nasr, S.M., & El Kholy, N.A., 2016. Effect of breed, apoVLDL-II gene polymorphism and metabolic biochemical markers on growth and body composition traits in commercial broiler breeds. Asian J. Anim. Vet. Adv. 11, 9, 548-555. https://doi.org/10.3923/ajava.2016.548.555 [ Links ]
Hellebrekers, L.J., Baumans, V., Bertens, A.P., & Hartman, W., 1990. On the use of T61 for euthanasia of domestic and laboratory animals: An ethical evaluation. Lab. Anim. 24, 200-204. https://doi.org/10.1258/002367790780866254 [ Links ]
Hirose, K., Ito, T., Fukawa, K., Arakawa, A., Mikawa, S., Hayashi, Y., & Tanaka, K., 2014. Evaluation of effects of multiple candidate genes (LEP, LEPR, MC4R, PIK3C3, and VRTN) on production traits in Duroc pigs. Anim. Sci. J. 85, 3, 198-206. https://doi.org/10.1111/asj.12134 [ Links ]
Houston, R.D., Cameron, N.D., & Rance, K.A., 2004. A melanocortin-4 receptor (MC4R) polymorphism is associated with performance traits in divergently selected Large White pig populations. Anim. Genet. 35, 5, 386-390. https://doi.org/10.1111/J.1365-2052.2004.01182.x [ Links ]
lyayi, E.A., & Odueso, O.M., 2003. Response of some metabolic and biochemical indices in rabbits fed varying levels of dietary cyanide. Afr. J. Biomed. Res. 6, 1, 43-47. https://doi.org/10.4314/ajbr.v6i1.54022 [ Links ]
Jiang, M.S., Chen, S.Y., Lai, S.J., Deng, X.S., Chen, Y., & Wan, J. (2008). Association between single nucleotide polymorphism of MC4R gene and carcass traits in rabbits. Hereditas (Beijing), 30, 12, 1574-1578 In Chinese. https://doi.org/10.3724/SP.J.1005.2008.01574 [ Links ]
Khalil, M.H., 2011. Rabbit genetic resources of Egypt. Anim. Genet. Resour. Inf. 26, 95-111. https://doi.org/10.1017/S101423390000122X [ Links ]
Khalil, M.H., Al-Sobayil, K., Hermes, I.H., & Al-Homidan, A.H., 2002. Crossbreeding effects for post-weaning growth, rectal and ear temperatures and respiration rates in crossing Saudi Gabali with Spanish V-Line rabbits. In: Proceedings.7th World Congress on Genetics Applied to Livestock Production, 19-23 August, Montpellier, France, pp. 4-12. [ Links ]
Lebas, F., 2000. Vitamins in rabbit nutrition: Literature review and recommendations. World Rabbit Sci. 8, 4, 185-192. https://doi.org/10.4995/wrs.2000.438 [ Links ]
Li, C., & Li, H., 2006. Association of MC4R gene polymorphisms with growth and body composition traits in chicken. Asian-Aust. J. Anim. Sci. 19, 6, 763-768. https://doi.org/10.5713/ajas.2006.763 [ Links ]
Martins, C., Cullere, M., Dalle Zotte, A., Cardoso, C., Alves, S.P., Bessa, R.J.B., Freire, J.P.B., & Falcão-e-Cunha, L., 2018. Incorporation of two levels of black soldier fly (Hermetia illucens L.) larvae fat or extruded linseed in diets of growing rabbits: Effects on growth performance and diet digestibility. Czech. J. Anim. Sci. 63, 9, 356-362. https://doi.org/10.17221/22/2018-CJAS [ Links ]
Medgyesi, G.A., Füst, G., Bazin, H., Ujhelyi, E., & Gergely, J., 1975. Interaction of rat immunoglobulins with complement. Federation of European Biochemical Societies (FEBS Lett). Antibody structure and molecular immunology Amsterdam: North-Holland; New York: American Elsvier, 36, PP 123-128. NLM ID: 101115835 [ Links ]
Meidtner, K., Wermter, A.K., Hinney, A., Remschmidt, H., Hebebrand, J., & Fries, R., 2006. Association of the melanocortin 4 receptor with feed intake and daily gain in F2 Mangalitsa χ Piétrain pigs. Anim. Genet. 37, 3, 245-247. https://doi.org/10.1111/j.1365-2052.2006.01414.x [ Links ]
Migdal, L., Palka, S., Kmiecik, M., & Derewicka, O., 2019. Association of polymorphisms in the GH and GHR genes with growth and carcass traits in rabbits (Oryctolagus cuniculus). Czech J. Anim. Sci. 64, 6, 255-264. https://doi.org/10.17221/27/2019-CJAS [ Links ]
Nahácky, J., Zidek, R., Gábor, M., Miluchová, M., Bucko, O., & Kovácik, A., 2018. MC4R and PGAM2 genes polymorphism association with production traits in rabbit (Oryctolagus cuniculus). J Microbiol. Biotech. Food Sci. 7, 5, 493-495. https://doi.org/10.15414/jmbfs.2018.7.5.493-495 [ Links ]
National Research Council, N.R.C., 1977. Nutrient Requirements of Rabbits. Second Revised Edition, Washington, DC: The National Academies Press. SBN: ISBN 0-309-02607-5 [ Links ]
Osaiyuwu, O.H., Bolaji, U.F.O., Adeyinka, O.A., Akinyemi, M.O. & Salako, A.E., 2020. Melanocortin 4 receptor (Mc4r) gene polymorphism and its association with body weights of some breeds of rabbit. Nig. J. Anim. Prod. 47, 2, 1 - 7. https://doi.org/10.51791/njap.v47i2.48 [ Links ]
Piles, M., Gomez, E.A., Rafel, O., Ramon, J., & Blasco, A., 2004. Elliptical selection experiment for the estimation of genetic parameters of the growth rate and feed conversion ratio in rabbits. J. Anim. Sci. 82, 3, 654-660. https://doi.org/10.2527/2004.823654x [ Links ]
Piórkowska, K., Tyra, M., Rogoz, M., Ropka-Molik, K.,Oczkowicz, M., & Rózycki, M., 2010. Association of the melanocortin-4 receptor (MC4R) with feed intake, growth, fatness and carcass composition in pigs raised in Poland. Meat Sci. 85, 2, 297-301. https://doi.org/10.1016/j.meatsci.2010.01 [ Links ]
Pla, M., Pascual, M., & Ariño, B., 2004. Protein, fat and moisture content of retail cuts of rabbit meat evaluated with the NIRS methodology. World Rabbit Sci. 12, 3, 149-158. https://doi.org/10.4995/wrs.2004.574 [ Links ]
Ramirez, R.M., Mota-Rojas, D., Reyes, A.D.L., Becerril-Herrera, M., Flores-Pintado, S., Alonso-Spilsbury, M., Cardona, L.A., Ramírez-Necoechea, R., & Lemus-Flores, C., 2006. Slaughtering process, carcass yield and cutting process in California and Chinchilla rabbit breeds. J. Food Technol. 4, 1, 86-89. https://www.researchgate.net/publication/282506075_Slaughtering_Process_Carc [ Links ]
Rochambeau, H. de., Duzert, R., & Tudela, F., 1998. Long term selection experiments in rabbit. Estimation of genetic progress on litter size at weaning. In Proc.: 6th World Congress on Genetics Applied to Livestock Production, Armidale, NSW, Australia, 11-16 January 1998, Vol. 26, 112-115. [ Links ]
Sanger, F., Nicklen, S., & Coulson, A.R., 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 12, 5463-5467. https://doi.org/10.1073/pnas.74.12.5463 [ Links ]
Shan, H., Song Z., Cao, Y., Xiong, P., Wu, J., Jiang, J., & Jiang, Y., 2020. Association of the melanocortin 4 receptor (MC4R) gene polymorphism with growth traits of Hu sheep. Small Rumin. Res. 192, 106206. https://doi.org/10.1016/j.smallrumres.2020.106206 [ Links ]
Song, X.M., Jiang, J.F., Zhang, G.Z., Shi, F.X., & Jiang, Y.Q., 2012. DNA polymorphisms of the Hu sheep melanocortin-4 receptor (MC4R) gene associated with birth weight and 45d-weaning weight. G.M.R. 11, 4, 4432-4441. http://dx.doi.org/10.4238/2012.September.27.3 [ Links ]
SPSS, 2015. IBM Corp. Released 2015. IBM SPSS Statistics for Windows, Version 23.0. Armonk, NY: IBM Corp. [ Links ]
Sternstein, I., Reissmann, M., Maj, D., Bieniek, J., & Brockmann, G.A., 2014. A new single nucleotide polymorphism in the rabbit (Oryctolagus cuniculus) myostatin (MSTN) gene is associated with carcass composition traits. Anim. Genet. 45, 4, 596-599. https://doi.org/10.1111/age.12165 [ Links ]
Szendrõ, Z., Matics, Z., Gerencsér, Z., Nagy, I., Lengyel, M., Horn, P., & Dalle Zotte, A., 2010. Effect of dam and sire genotypes on productive and carcass traits of rabbits. J. Anim. Sci. 88, 2, 533-543. https://doi.org/10.2527/jas.2009-2045 [ Links ]
Tamura, K., Dudley, J., Nei, M., & Kumar, S., 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, software version 4.0, 24, 8, 1596-1599.https://doi.org/10.1093/molbev/msm092 [ Links ]
Testik, A., 1996. The situation of rabbit production and production performances of some exotic rabbits in Turkey. In: Proc. 6 th World Rabbit Congress, 1996 July, Toulouse, France, Vol. 3, 435-436. [ Links ]
Tian, B., 2005. Identification of direct genomic targets downstream of the nuclear factor-kappa B transcription factor mediating tumor necrosis factor signaling. J. Biol. Chem. 280, 17, 17435-17448. https://doi.org/10.1074/jbc.M500437200 [ Links ]
Trocino, A., Cotozzolo, E., Zomeño, C., Petracci, M., Xiccato, G., & Castellini, C., 2019. Rabbit production and science: the world and Italian scenarios from 1998 to 2018. Ital. J. Anim. Sci. 18, 1, 1361-1371. https://doi.org/10.1080/1828051X.2019.1662739 [ Links ]
Wagner, D.D., Furrow, R.D., & Bradley, B.D., 1983. Subchronic toxicity of monensin in broiler chickens. Vet. Pathol. 20, 3, 353-9. https://doi.org/10.1177/030098588302000311 [ Links ]
Wang, Y., Su, Y., Jiang, X.S., & Liu, Y.P., 2009. Study on association of single nucleotide polymorphism of MC3R and MC4R genes with carcass and meat quality traits in chicken. J. Poult. Sci. 46, 3, 180-187. https://doi.org/10.2141/jpsa.46.180 [ Links ]
Wardlaw, S.L., 2001. Obesity as a neuroendocrine disease: Lessons to be learned from proopiomelanocortin and melanocortin receptor mutations in mice and men. J. Clin. Endocrinol. Metab. 86, 4, 1442-1446. https://doi.org/10.1210/jcem.86.47388 [ Links ]
Xuemei, Q., Ning, L., Xuemei, D., Xingbo, Z., Qingyong, M. & Xiuli, W., 2006. The single nucleotide polymorphisms of chicken melanocortin-4 receptor (MC4R) gene and their association analysis with carcass traits. Science in China Series C: Life Sci. 49, 6, 560-566. https://doi.org/10.1007/s11427-006-2029-7 [ Links ]
Yamani, K. A., 1994. Rabbit meat production in Egypt. 1st International Conference of rabbit production in hot climates, In: Baselga M. (ed.), Marai I.F.M. (ed.). Rabbit production in hot climates. Zaragoza, 1994/09/06-08, Cairo (Egypt), CIHEAM - Cah. Options Méditerr. 8, 57-64. http://om.ciheam.org/om/pdf/c08/95605279.pdf [ Links ]
Zaghloul, A.R., Khalil, M.H., Iraqi, M.M., Ramadan, Sh., & EL Nagar, A.G., 2019. Crossbreeding effects and polymorphic associations of genotypes of GH gene with growth traits in rabbits. EJRS 29, 2, 235-255. https://doi.org/10.21608/EJRS.2019.81100 [ Links ]
Zhang, C.L., Wang, Y.H., Chen, H., Lan, X.V., Lei, C.Z., & Fang, X.T., 2009. Association between variants in the 5'- untranslated region of the bovine MC4R gene and two growth traits in Nanyang cattle. Mol. Biol. Rep. 36, 7, 1839-1843. https://doi.org/10.1007/s11033-008-9388-z [ Links ]
Zhang, G.W., Gao, L., Chen, S.Y., Zhao, X.B., Tian, Y.F., Wang, X., Deng, X.S., & Lai, S.J., 2013. Single nucleotide polymorphisms in the FTO gene and their association with growth and meat quality traits in rabbits. Gene 527, 2, 553-557. https://doi.org/10.1016/j.gene.2013.06.024 [ Links ]
Zhang, W.X., Zhang, G.W., Peng, J., & Lai, S.J., 2012. The polymorphism of GHR gene associated with the growth and carcass traits in three rabbit breeds. In: World Rabbit Science Association. Proceedings of the 10th World Rabbit Congress, September 3 - 6, 2012, Sharm El-Sheikh, Egypt, 75-78. [ Links ]
Zhou, Y., Cao, D.G., Lei, Q.X., Han, H.X., Li, F.W., Li, G.M., & Huang, B.H., 2012. Associations of melanocortin-4 receptor (MC4R) gene single nucleotide polymorphisms with carcass traits in a synthetic broiler line. J. Anim. Vet. Adv. 11, 1, 13-19. https://doi.org/10.3923/javaa.2012.13.19 [ Links ]
Submitted 9 May 2022
Accepted 22 August 2022
Published January 2023
# Corresponding author: hend_radwan@mans.edu.eg














