Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Animal Science
versão On-line ISSN 2221-4062
versão impressa ISSN 0375-1589
S. Afr. j. anim. sci. vol.52 no.5 Pretoria 2022
http://dx.doi.org/10.4314/sajas.v52i5.02
The effect of different feeding strategies on honey bee gut microbiota and the presence of Nosema
E. TopalI; Ö. CeylanII; R. I. TuncaIII, #; V. BayIV; S. AldemirV; H. inciVI; U. TopçuogluV; M. KösogluV
IIzmir Food Control Laboratory Directorate, Bornova, Izmir, Türkiye
IIDept. of Food Processing, Ula Ali Kocman Vocational School, Mugla Sitki Kocman University, Mugla, Türkiye
IIIDept. of Plant and Animal Breeding, Ula Ali Kocman Vocational School, Mugla Sitki Kocman University, Mugla, Türkiye
IVDepartment of Animal Science, Faculty of Agriculture, Ege University, Izmir 35100, Türkiye
VAegean Agricultural Research Institute, Izmir 35661, Türkiye
VIDepartment of Animal Science, Faculty of Agriculture, Bingöl University, Bingöl 12000, Türkiye
ABSTRACT
Colony development in honey bees depends on both the environmental conditions and their genetic structure. Nutrition is the one of the most important factors in the honey bee's health. This study was carried out in 48 colonies and included six groups (a control and five feeding groups). In the experiment, the effect of different feeding strategies on intestinal flora and the presence of Nosema was investigated. Pantoea agglomerans, Pseudomonas luteola, Burkholderia cepacia, Brevibacillus nitrificans, Sphingomonas paucimobilis, Aeromonas hydrophila, and P. alcalidimonas were detected in the intestinal microflora of the bee samples by morphological and phenotypic identification. The results of phenotypic identification were confirmed using 16S rRNA sequence analysis for P. agglomerans and B. nitrificans strains. The presence of Nosema was simultaneously investigated in all groups. Only Nosema cerenae was detected using DNA analysis in the positive Nosema spore samples.
Keywords: Apis mellifera anatolica, feeding, Nosema, intestinal flora
Introduction
Beekeeping is a production activity that depends on environmental conditions. In recent years, a topic that has received attention (after the colony loss studies) is the intestinal microflora of honey bees. It has been understood that the intestinal microflora of the honeybee play a role in the metabolism, immune function, growth, and development of the bee, and are especially effective in the preservation of bee health. Chemicals used in the fight against parasites and pathogens, season, flora, food sources, age of the individual or job in the hive, and many other factors can affect the intestinal microflora of honey bees (Aldemir et al., 2019; Castelli et al., 2020; Papp et al., 2021; Marín-García et al., 2022; Ke et al., 2022). Microorganisms in the intestines of honeybee not only aid in the digestion of food, but also contribute to detoxification, protection against pathogens and parasites, modulation enhancement, and immunity (Flint et al., 2012; Hooper et al., 2012; Engel & Moran, 2013; Maes et al., 2016; Attia et al, 2019; Dimov 2022; Ricigliano et al., 2022). The intestinal microflora of honey bees consists mostly of facultative anaerobic and micro-aerophilic bacteria adapted to the host. Overall, developments including reports of extreme variation, increased protective and nutritive functions, and disease prevalence have necessitated the study of intestinal symbionts and microflora as a potential model in bee health. Bee intestinal microflora are dominated by only nine bacterial species (Bartonella apis, Parasaccharibacter apium, Frischella perrara, Snodgrassella alvi, Gilliamella apicola, Bifidobacterium spp., Lactobacillus Form-4, Lactobacillus Form-5, and Other) and are a community that is much simpler than the mammalian microbiota (Kwong & Moran, 2016). The gut microbiota of the honey bee provides several advantages as an experimental system for investigating how gut microbiota communities affect their hosts and the processes that determine gut community composition and dynamics. To date, studies of the honeybee gut microbiota show that it influences the host. In particular, nutrition, weight gain, endocrine system, immune function, and pathogen resistance have caused changes in the health status of host bees with changes in microbiota (Zheng et al., 2018).
The most important causes of deterioration in honey bee health are nutritional deficiency, parasites, and pathogens (Dolezal & Toth, 2018). During the honey flow period, Nosema and microsporidian infections mostly affect field bees. It has been reported that pollen and bee bread can be a source of spores (Sokol & Michalczyk, 2016). The quality and diversity of pollen positively affects bee health and longevity (Di Pasquale et al., 2013). Nosema is associated with stress and increases with nutrient deficiencies and manipulation in rearing. High Nosema levels occur during breeding as a result of protein deficiency in colonies in late autumn and winter. The autumn period is an important time of the year for hive management to prevent spring outbreaks of Nosema disease. Nosema apis affects the wintering performance of adult bees. In temperate regions, Nosema apis infections should be seen as a serious problem and their adverse effects on the productive capacity of honeybee colonies in these regions should not be ignored (Fries, 1993; Rice, 2001; Somerville, 2005).
In generally, parasites alter the foraging behavior of their hosts to make changes in feed intake rates. Studies conducted on Nosema ceranae in European or Western honey bees have reported that these bees increase their energy needs. Infected bees fed high-quality pollen were compared to infected bees fed lower quality pollen or a pollen-free diet. Bees fed high-quality pollen were more likely to survive (Ferguson et al., 2018). Honey bees not infected with Nosema show no difference in survival on diets of different pollen qualities. According to the study, it was determined that bees infected with N. ceranae benefit from the increase in pollen quality and prefer higher quality pollen when foraging for pollen, but the infection did not affect pollen collection at the individual or hive levels (Ferguson et al., 2018).
Pollen is the main source of protein and lipids for honey bees. Especially during the winter period, nutritional differentiation poses a threat to the continuity of the hives. Therefore, in this study, changes in the intestinal microflora of honey bees and the presence of Nosema in colonies fed with different pollens before wintering were determined.
Material and Methods
The study was carried out in the apiary of the Aegean Agricultural Research Institute of Menemen District of Izmir Province in Türkiye (38°33'54" N; 27°3'27" E). In the experiment, sister queens of the Efe Bee (Apis mellifera anatoliaca), which were registered as a result of the breeding work carried out by the Aegean Agricultural Research Institute in 2020 were used. Colonies were formed on 14 September 2020 from three frames and a 1-kg package of bees. Experimental groups were composed of eight colonies in each group with six groups, including the control, syrup, Rocky rose (Cistus creticus) pollen, Poppy (Papaver somniferum L.) pollen, mixed pollen, and the commercial bee cake group, giving a total of 48 colonies.
In the study, poppy pollen, an industrial plant, rocky rose, mixed spring pollen, and sugar syrup made from beet sugar was used as a natural flora sources. Pollen from producers was stored in a deep freeze (-20 °C) until use. Commercial bee cakes were selected from pollen-added products sold in the market. While fresh pollen was used for the protein requirement of the colony, a beet sugar-water mixture was used to meet the carbohydrate needs of the colony. In order to ensure the freshness of the pollen and to observe the consumption and storage rate in the colonies, it was provided to the colonies according to the consumption status. The colony was fed with a 2:1 sugar-water mixture for the formation of honey stores. The group formed as the control group was given 1 liter of syrup in order to eliminate the stress of the first day. The first feeding was carried out on the first day of the creation of the trial material. The research was planned according to a random plots trial design. Honey bee samples were collected in autumn, 2020 (November) and early spring, 2021 (March) after wintering in order to determine the effects of feeding on colony performance between groups.
Intestinal microbiota: In the experiment, a total of 30 bee samples (15 forager and 15 nurse bees) were collected from each hive. The live bees removed from the containers were kept in a 4 °C refrigerator for 4 min to reduce their motility. After this process, each bee sample was coded as a nurse or forager, with the feeding element, and then the abdominal segments and intestinal contents of the bees were removed. For the determination of intestinal microflora, eight different samples for each of the different feeding groups and 96 different samples as a nurse-forager were sampled at two different times, giving a total of 198 different intestinal samples, which were spread on individual petri dishes.
The obtained intestinal contents were inoculated as three samples on yeast-malt agar (YMA; Difco) medium containing two different additives. By adding 100 mg/mL ampicillin to this medium, bacterial growth was prevented; by adding 100 mg/mL cycloheximide, yeast growth was inhibited, and bacterial growth was encouraged (Good et al., 2014). The petri dishes were incubated in an aerobic environment at 25 °C for 3-5 days for different time periods. From the bacterial colonies grown in the petri dishes, those with different morphological structures were taken into separate petri dishes using sterile cultivation techniques. They were classified according to their common colony morphotypes. The pure bacteria obtained were classified using the Gram staining method and API 20 NE test kits were used for species identification of gram-negative, non-enteric Bacillus forms (Figure 1). Species identification was performed using APIWEB software, which is a 7-digit number formed according to the 24- and 48-hour results of different phenotypic characters available in the test kits for each different isolate. All the obtained pure bacterial samples were analysed in three replicates.
In the study, 16S rRNA sequence analyzes were performed by a commercial company (BM Labosis) in order to confirm the phenotypic identification results. Four isolates were sent for molecular identification. The EurX GeneMATRIX Bacterial and Yeast DNA isolation kit was used for DNA isolation. In the PCR study, gene regions targeted for species identification were amplified with 27F-1492R primers as universal primers. The primer sequences were: 27F 5' AGAGTTTGATCMTGGCTCAG 3'; 1492R 5' TACGGYTACCTTGTTACGACTT 3'. The PCR product was purified with the MAGBIO "HighPrep PCR Clean-up System". Reads obtained with primers 27F and 1492R were configured in BioEdit software and the species was determined according to the closest species on BLASTN (NCBI).
Honey bee samples were taken in November, 2020. In the experiment, 50 forager worker bees per apiary, representing each hive, were collected and brought to the laboratory and stored in the freezer until analysis. The method specified in the World Organization for Animal Health (OIE) Application Guide (2008) was used for the homogenates prepared to determine the level of Nosema. Nosema spore counts from homogenates were performed on a haemocytometer using a light microscope (OIE, 2008). DNA isolation was performed on positive samples with a detected Nosema spore level using a commercial DNA isolation kit (Thermo Genomic DNA Purification Kit, K0722).
The total PCR reaction volume was 25 μΙ: 5 μΙ of DNA was prepared using 2X DreamTaq DNA Polymerase Master Mix, 10 pmol of 218MITOC FOR and REV, 10 pmol of μM 321APIS FOR and 321APIS REV primers, and UP H2O. Special primers for N. apis and N. ceranae were used in the PCR process (Table 1).
PCR conditions were 2 min at 95 °C, 35 cycles after initial denaturation; 45 s at 95 °C; 45 s at 59.3 °C; 1 min at 72 °C; and 7 min at 72 °C, following the last cycle, with storage at 4 °C. PCR products were run on 1.2% agarose gel and stained with GelDYE and visualized under ultraviolet light. The sequence data obtained after sequencing the PCR products were determined using the BLAST program on NCBI to determine the types of Nosema spores. After the Nosema spore data were entered into the IBM SPSS (2021) program, a normality test was applied to the samples. Since the data were not normally distributed, the samples were subjected to the nonparametric Kruskal-Wallis test and statistically significant groups are reported.
Results
The climate data during the trial period are given in Table 2. In particular, the winter period was not as cold as previous years.
Differences in gut microbiota between feeding groups: Different colonies of microorganisms grown in each inoculated petri dish were classified according to their morphological differences. Some petri images are shown in Figure 2 as an example of the different microorganism colonies growing in isolation.
Colony morphology and structure, colony colour, and colony number of the microorganisms grown in each inoculated petri dish were determined separately for microorganisms and yeasts (Table 3). Table 3 represents the samples taken in the fall of 2020 and the samples taken in the early spring of 2021.
A total of 91 different strains with different colony morphology and structure were obtained from the isolated petri dishes. The purification cultivations were made for each of these strains (Figure 3). These different strains were stocked in petri dishes and flat, NA petri dishes.
When the isolation petri dishes were examined, the differences were determined in the total microorganism and total yeast loads according to the feeding groups. It was determined that the total microorganism load of the bees fed with mixed pollen, commercial bee cake, and syrup was higher than the rocky rose (Cistus creticus), poppy (Papaversomniferum L.), and control groups. Especially in honey bees fed with mixed pollen, commercial bee cake, and syrup, an increase in microorganism load was observed in nurse bees over time. Honey bees fed rocky rose and poppy caused a slight rise in the microorganism load.
It was determined that the highest microorganism development in field bees was in the honey bee samples fed with poppy pollen in the autumn period of 2020. However, it was observed that the microorganism load decreased in the early spring (2021) poppy pollen group. The same results were also seen in field bee samples fed with mixed pollen. Microorganism load decreased in the early spring group of 2021 in bees that continued to be fed with mixed pollen. Field bees fed with commercial bee cake and syrup showed an increase in intestinal microbial load over time. Compared to the control group fed with rocky rose pollen, it was observed that there was no change in the intestinal microflora of the bees and field bees.
A reduction in yeast load was observed in all feeding groups for both keepers and field bees. While a higher number of yeasts were detected in the bee intestinal microflora at the start of feeding, a decrease in yeast load was detected in all other feeding groups, except syrup feeding in nurse bees.
Identification results with API 20 NE test kits showed that there were changes in microorganism species according to nutritional groups (Table 4). The most intensely detected species were P. agglomerans and P. luteola species. Pantoea agglomerans was detected in the intestinal microflora of all nutritional groups. Pseudomonas luteola was observed in the intestinal microflora of all feeding groups except honey bees fed with sugar syrup. These two species, and S.paucimobilis, B. nitrificans and A. hydrophila species were detected in bee samples fed with rocky rose pollen. In the intestinal microflora of honey bees fed with poppy pollen, P. agglomerans, P.luteola, and B. nitrificans, B. cepacia and P. alcaligenes species were determined. Pantoea agglomerans and P. luteola as well as B. cepacia species were observed in bee samples fed with the commercial bee cake. In the intestinal microflora of the bee samples fed only with syrup, besides P. agglomerans and B. vesicularis, A. hydrophila species were isolated.
The phenotypic identification and the 16S rRNA sequence analysis results are given in Table 5.
Nosema differences between the feeding groups: Nosema spore numbers of forager and nurse bees in the groups are given in Figure 4. Groups are indicated as A (commercial bee cake), C (control), H (poppy pollen), K (mixed pollen), P (rocky rose pollen) and S (syrup).
A difference was found in the level of Nosema spores between forager bees fed syrup and rocky rose pollen (p <0.05). No statistically significant relationship was found between the Nosems level in nurse bees (Fig. 4).
When all bees (forager and nurse bees) were evaluated together, a difference was found in Nosema levels between syrup and rocky rose groups (p <0.01) (Fig. 5).
No statistically significant difference was found between the groups in the level of Nosema in forager/nurse bees (Figure 6, Figure 7).
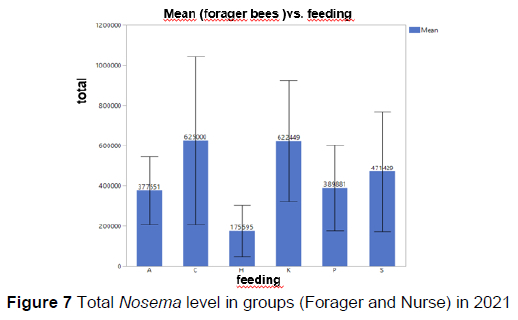
There was a seasonal difference in Nosema level between field bees in the commercial bee cake and rocky rose groups (p <0.05), and between forager bees and total spore number in the syrup group. However, the seasonal difference in the Nosema level in the control, poppy pollen, and mixed pollen groups was not significant. Nosema ceranae was detected using DNA sequencing of positive samples. Nosema ceranae sequences were similar to those reported in China, Mexico, Poland, Iran, Lithuania, Italy, Saudi Arabia, and Argentina after BLAST analysis.
Discussion
According to the results of the study, the total intestinal microorganism load increased in the nurse bees in different feeding groups. However, the microbial load was only increased in forager bees fed with commercial bee cake and syrup. The total yeast load tended to decrease in the feeding groups. The present study results are in accordance with another study, which reported that total bacterial loads differed significantly among foragers, caretaker, and winter bees (Kesnerovà et al., 2020). According to their result, long-lived winter bees have the highest bacterial load and the lowest community α-diversity (Kesnerovà et al., 2020). In another study examining the composition of the intestinal bacteriota of honey bees, a significant difference was found between the samples taken in March and May. A significant difference in composition was reported between the colder and warmer regions in the intestinal contents of the March samples (Papp et al., 2021). In another study, it was determined that the diversity of the general gut microbiota varied significantly according to the type of diet (Huang et al., 2018). It has been reported that N. ceranae infection substantially affects the intestinal microbiota in bees fed only syrup. Lactobacillus, Gluconacetobacter, and Snodgrassella were higher in bees fed with bee bread than those fed with sugar water but Serratia was detected in lower numbers in that study. Both feeding types and N. ceranae infection significantly affect the gut microbiota of A. cerana foragers. It has been reported that bee bread, which is a good food source, creates a more stable intestinal microbiota in honey bees and therefore protects bees against N. ceranae infection (Huang et al., 2018).
In our study, mixed pollen was given, considering that it is a more complicated food source. However, since the growth rate of this group of colonies was not at the desired level, it is thought that the pollen may have been old. In a study supporting this theory, development was impaired and the mortality rate increased in bees fed with old diets. It has been reported that changes in diet quality play an important role in colony health and the establishment of a dysbiotic gut microbiome (Maes et al., 2016).
Colony health is closely related to the bacteria in the gut of honeybees. The type of sugar used in the feed during the winter months appeared to affect the relative abundance of the dominant bacterial communities in the guts of their colonies. The presence of supernal Alphaproteobacteria (Acetobacteraceae), Bifidobacterium, and Lactobacillaceae in the gut of sucrose-fed honey bees has been reported to be a very suitable for honeybees during winter (Wang et al., 2020).
As a result of our feeding studies, an increase in the microbial load of the nurse bees and a decrease in the yeast load of both the nurse and field bees were observed. This study is one of the first studies to determine the intestinal microbial flora of honey bees for Turkey and the Efe bee. In our study, Pantoea agglomerans, Pseudomonas luteola, Brevibacillus nitrificans, Sphingomonas paucimobilis, Aeromonas hydrophila, Burkholderia cepacia, Pseudomonas alcaligenes, and Brevundimonas vesicularis species were determined using morphological identification kits in nurse and forager honeybees to indicate a change in microbial load occurring in the intestinal flora of different feeding groups. In a study conducted in Ordu province, the presence of species belonging to Staphylococcus, Klebsiella, Citrobacter, Leuconostoc, Kocuria, Sphingomonas, Burkholderia, Hafnia, Escherichia, Aeromonas, Pantoea, Bacillus, Paenibacillus, and Streptococcus genera were reported in the microbial flora of healthy and dead adult bees (Yarilgaç, 2016). The study supported that Escherichia coli and Bacillus licheniformis had the most lethal effect on bees and indicated a microbial load in support of our study (Yarilgaç, 2016).
In our study, while there was no statistical difference between the groups in the level of Nosema between the nurse bees, a difference was found between the syrup and rocky rose pollen groups in foragers (p <0.05). It has been observed that nutrition and quality affect the Nosema level. In a study supporting our findings, it has been reported that feeding honey bees in cages in the laboratory with polyfloral pollen increases immune-related enzyme activities, making them more resistant to stress. It has been reported that feeding the colonies with good pollen increases the resistance of the honey bee to the destroyer, N. ceranae, or the ectoparasitic mite, Varroa (Huang, 2012). In another study, honey bees were susceptible to many diseases, including Nosema, which can decrease population size of the colony during the winter months. It has been reported that Fe, Mn, Ni, and Na deficiencies observed in Nosema-infected bees may be the cause of more deaths in these colonies during the wintering period. There is a strong correlation between the bioelement content in honey bees and season, and Nosema infection (Ptaszynska et al., 2018). In addition to malnutrition, honey bees face overlapping honey bee diseases and various environmental challenges. Azzouz-Olden et al. (2018) studied the effects of feeding natural and artificial rations on the abdomen of the honey bee and the relationship of these effects with Nosema was investigated. It was determined that pollen feeding improved feeding behavior, positively affected the hunger gene, and was effective against Nosema (Azzouz-Olden et al., 2018).
In our study, after the wintering period in the groups; one colony in the commercial bee cake group, three colonies in the syrup group, one colony in the mixed pollen group, and four colonies in the control group were killed. It can be said that the poor food quality, disease, and pest status have an effect on the life of the honey bee.
In another study that supports our results, it was determined that infected bees fed a high-quality pollen were more likely to survive than Nosema-infected bees fed a lower quality pollen or no pollen. Bees infected with N. ceranae prefer higher quality pollen when foraging, but the infection status does not change the amount of pollen collected by the bee or the colony (Ferguson et al., 2018). Another study reported that Eucalyptus grandis pollen promoted the proliferation of Nosema ceranae (Castelli et al., 2020).
Nosema infestation is affected by the season as well as nutritional quality. In another study supporting our finding, it was determined that the intensity of Nosema infections in honey bee colonies changed seasonally during the year, with the highest spore numbers of the parasite observed in the spring, and the lowest in the autumn and winter periods (Traver et al., 2012). In another study performed in the USA, the highest prevalence and spore viability rates of N. ceranae infection were found in spring and summer, while the lowest rates were found in autumn. It was determined that N. ceranae spore viability was significantly related to the prevalence and infection density in bees. A high N. ceranae infection (>1,000,000 spores/bee) has been reported to be related to the decreasing bee populations and food stores in the colonies, and that treatment is absolutely necessary at this level of spores. In addition, the survival rate of worker bees is significantly reduced due to N. ceranae infections. It has been reported that N. ceranae infections could be harmful for colony productivity (Emsen et al., 2020).
In a study performed in the Black Sea region, the total infection rate in worker bees was determined as 21.23%. It has been reported that the infection rate of N. ceranae increases proportionally with an increase in temperature and humidity factors. Relative humidity is more conducive than temperature on the N. ceranae infection rate. In addition, the highest infection rates were observed in June and July; the N. ceranae infection rate was found to be higher in low-altitude regions (Tosun & Yaman, 2016).
Studies on the determination of Nosema species in bee samples using molecular methods have been carried out by some researchers (Whittekar et al., 2010). Molecular identification of N. ceranae has been done in Turkey (Ütük et al., 2010). In another study, 89% N. ceranae and 11% N. apis infections were detected in the colonies in the Hatay wintering region, while 84% N. ceranae and 16% N. apis infections were detected in the samples from the southeast Marmara region (Muz et al., 2010).
In another study conducted in Turkey, 4640 dead adult worker bees collected from 20 locations in the Eastern Black Sea region were studied. They determined that 985 (21.23%) of 4640 samples were infected with N. ceranae using molecular techniques (Tosun & Yaman, 2016). The presence of Nosema spores at different rates has been determined in Kirsehir province and its districts. In all Nosema-spore-positive samples, N. ceranae was the prime disease agent in that region (Büyük et al., 2017).
In a study carried out in Mugla province in Turkey, Nosema spores were counted in samples taken from 152 apiaries located in 13 different locations and 62 water sources close to these apiaries, and the molecular diagnosis was made from samples with Nosema spores. In the network analysis, three haplotypes were determined for the first time according to these gene regions. The prevalence of Nosema disease in Mugla region was determined as 71.53 ± 6.02% and the presence of only N. ceranae was determined as the disease factor. In addition, BLAST analysis has been reported to have a high similarity (94-100%) with N. ceranae samples previously reported in Lebanon, France, Morocco, and Thailand (Kartal et al., 2021).
Our study also supports the information that the Nosema species isolated from bee samples are N. ceranae. In studies on the subject, it has been reported by many researchers that N. ceranae started to become widespread in Turkey and became dominant in the process (Tunca et al., 2016; Öziçli & Aydin, 2018; Saribiyik & Özkim, 2018).
According to a study investigating the interactions between the honey bee gut microbiome and N. ceranae, when experimentally infected bees sampled 5, 10, and 21 days after infection were examined, they detected variation in infection levels at the colony level. There are differences between the microbiota of colonies with high infection levels and colonies with low infection levels (Rubanov et al., 2019).
Conclusions
In the current study, intestinal microbial flora of nurse and forager bees were determined in autumn and early spring. The changes in the intestinal microflora of bees fed with rocky rose pollen, poppy pollen, mixed pollen, commercial bee cake, and syrup were investigated. An increase in intestinal microbial load was observed in nurse bees in all feeding groups used in contrast to the control group. However, in forager bees, feeding with poppy pollen and mixed pollen caused a decrease in intestinal microbial load, while feeding with commercial bee cake and syrup showed a positive contribution to microbial load. No changes were observed between the rocky rose pollen and the control group. In the intestinal microbial flora studies, it was determined that the yeast growth was at a very low level compared to the bacterial growth. Except for the nurse bees fed only with syrup, a decrease in the intestinal yeast loads of the nurse and forager bees was detected in all other feeding groups.
In this study, P. agglomerans and P. luteola species were determined as the dominant species among the gram-negative bacteria detected in the intestinal microflora of bee samples in all feeding groups. Sphingomonas paucimobilis, Aeromonas hydrophila, Burkholderia cepacia, Pseudomonas alcaligenes, Brevundimonas vesicularis species were identified. In order to confirm our phenotypic identification results, P. agglomerans and B. nitrificans strains were confirmed using 16S rRNA sequence analysis.
According to the results of this research performed with different food groups, it was found that the Nosema spore levels were statistically significant between commercial bee cake and cotton foragers, and between syrup group foragers, in total and also seasonally. The effects of nutritional differences in the Nosema spore level and the season were clearly revealed. The spores observed in both autumn and spring periods were only N. ceranae. These results show that N. ceranae has become dominant in our country.
Feeding with different food groups and seasonal differences may cause changes in bee intestinal flora and Nosema spore density. The results obtained are important in determining new feeding strategies for beekeepers. A well-fed colony can maintain colony resistance by strategies developed by the intestinal flora, even if the parasite pathogen density has increased in the colonies. Our study results are supported by other studies. Quality of nutrients is very important for the continuation of the colony.
Acknowledgements
This article was produced by the project "The Pollen Preferences of Honeybees and the Effects of Pollen Use in Winter on Colony Dynamics" (TAGEM/HAYSÜD/B/20/A4/P5/1890), supported by The Turkish Ministry of Agriculture and Forestry, General Directorate of Agricultural Research and Policies (TAGEM). We would like to acknowledge TAGEM and the Aegean Agricultural Research Institute for their support.
Authors' contributions
ET, MK, RÍT, and ÖC contributed to the project idea, design, and execution of the study. ET, MK, UT, and VB contributed to the acquisition of data. ET, RÍT, ÖC, VB, SA, and UT contributed to laboratory analyses. VB and ÖC analysed the data. ET, RÍT, ÖC, and SA drafted and wrote the manuscript. RÍT, ÖC, and HÍ reviewed the manuscript critically. All authors have read and approved the finalized manuscript.
Conflict of interest declaration
The authors declare that they have no conflict of interest.
References
Aldemir, S., Tunca, R.Í., Topal, E. & Margaoan, R., 2019. Bal arilarinin bagirsak yapisina etki eden faktörler. Aricilik Arastirma Dergisi, 11(1), 28-34. [ Links ]
Attia, Y.A., Bovera, F., Abd-Elhamid, A.E.H.E., Calabrò, S., Mandour, M.A., Al-Harthi, M.A. & Hassan, S.S., 2019. Evaluation of the carryover effect of antibiotic, bee pollen, and propolis on growth performance, carcass traits and splenic and hepatic histology of growing rabbits. J Anim Physiol Anim Nutr. 103(3), 947-958. [ Links ]
Azzouz-Olden, F., Hunt, A. & Degrandi-Hoffman, G., 2018. Transcriptional response of honey bee (Apis mellifera) to differential nutritional status and nosema infection. BMC Genomics, 19.1: 628. https://doi.org/10.1186/s12864-018-5007-0 [ Links ]
Büyük, M., Tunca. R.Í. & Taskin, A., 2017. Kirsehir ilindeki ariliklarda nosema hastaliginin belirlenmesi. Türk Tarim-Gida Bilim ve Teknoloji dergisi, 5(1), 1-5. [ Links ]
Castelli, L., Branchiccela, B., Garrido, M., Invernizzi, C., Porrini, M., Romero, H. & Antúnez, K., 2020. Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae infection. Microb. Ecol. 80(4), 908-919. https://doi.org/10.1007/s00248-020-01538-1 [ Links ]
Di Pasquale, G., Salignon, M., Le Conte, Y., Belzunces, L.P., Decourtye, A., Kretzschmar, A. & Alaux, C., 2013. Influence of pollen nutrition on honey bee health: Do pollen quality and diversity matter? PloS one, 8(8), e72016. https://doi.org/10.1371/journal.pone.0072016 [ Links ]
Dimov, S.G., 2022. Environmental and anthropogenic influence on the core beneficial honeybee gut microbiota-a short communication from Bulgaria. Bacteria, 1(2), 88-95. [ Links ]
Dolezal, A.G. & Toth, A.L., 2018. Feedbacks between nutrition and disease in honey bee health. Curr. Opin. Insect. Sci. 114-119. https://doi.org/10.1016/j.cois.2018.02.006 [ Links ]
Emsen, B., De la Mora, A., Lacey, B., Eccles, L., Kelly, P.G., Medina-Flores, C.A. & Guzman-Novoa, E., 2020. Seasonality of Nosema ceranae infections and their relationship with honey bee populations, food stores, and survivorship in a North American region. Vet Sci. 7(3), 131. https://doi.org/10.3390/vetsci7030131 [ Links ]
Engel, P. & Moran, N.A., 2013. The gut microbiota of insects-diversity in structure and function. FEMS Microb. Rev. 37(5), 699-735. https://doi.org/10.1111/1574-6976.12025 [ Links ]
Ferguson, J.A., Northfield, T.D. & Lach, L., 2018. Honey bee (Apis mellifera) pollen foraging reflects benefits dependent on individual infection status. Microb Ecol. 1-10. https://doi.org/10.1007/s00248-018-1147-7 [ Links ]
Flint, H.J., Scott, K.P., Louis, P. & Duncan, S.H., 2012. The role of the gut microbiota in nutrition and health. Nature Reviews Gastroenterology & Hepatology, 9(10), 577. https://doi.org/10.1038/nrgastro.2012.156 [ Links ]
Fries, I.,1993. Nosema apis: A parasite in the honey bee colony. Bee World. 74: 5-19. https://doi.org/10.1080/0005772X.1993.11099149 [ Links ]
Good, A.P., Gauthier, M.P.L., Vannette, R.L. & Fukami, T., 2014. Honey bees avoid nectar colonized by three bacterial species, but not by a yeast species, isolated from the bee gut. PLoS One, 9(1), e86494. https://doi.org/10.1371/journal.pone.0086494 [ Links ]
Hooper, L.V., Littman, D.R., & Macpherson, A.J., 2012. Interactions between the microbiota and the immune system. Science 336, 1268-1273. DOI: 10.1126/science.1223490 [ Links ]
Huang, Z., 2012. Pollen nutrition affects honey bee stress resistance. Terr Arthropod Rev. 5(2), 175-189. https://doi.org/10.1163/187498312X639568 [ Links ]
Huang, S.K., Ye, K.T., Huang, W.F., Ying, B.H., Su, X., Lin, L.H. & Hu, J.Z., 2018. Influence of feeding type and Nosema ceranae infection on the gut microbiota of Apis cerana workers. MSystems, 3(6), e00177-18. https://doi.org/10.1128/mSystems.00177-18. [ Links ]
Kartal, S., Tunca, R.Í., Özgül, O., Karabag, K. &, Koç, H., 2021. Microscopic and molecular detection of Nosema sp. in the southwest Aegean Region. Uludag Aricilik Dergisi, 21 (1), 8-20. https://doi.org/10.31467/uluaricilik.880380 [ Links ]
Kesnerová, L., Emery, O., Troilo, M., Liberti, J., Erkosar, B., & Engel, P., 2020. Gut microbiota structure differs between honeybees in winter and summer. The ISME Journal, 74(3), 801-814. https://doi.org/10.1038/s41396-019-0568-8 [ Links ]
Ke, L., Yan, W.Y., Zhang, L.Z., Zeng, Z.J., Evans, J.D. & Huang, Q., 2022. Honey bee habitat sharing enhances gene flow of the parasite Nosema ceranae. Microb Ecol. 83(4), 1105-1111. [ Links ]
Kwong, W.K. & Moran, N.A., 2016. Gut microbial communities of social bees. Nature Reviews Microbiology, 74 (6), 374. https://doi.org/10.1038/nrmicro.2016.43 [ Links ]
Maes, P.W., Rodrigues, P.A., Oliver, R., Mott, B.M., & Anderson, K.E., 2016. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol Ecol., 25(21), 5439-5450. [ Links ]
Marín-García, P. J., Peyre, Y., Ahuir-Baraja, A.E., Garijo, M.M. & Llobat, L., 2022. The role of Nosema ceranae (Microsporidia: Nosematidae) in honey bee colony losses and current insights on treatment. Vet Sci. 9(3), 130. [ Links ]
Muz, M.N., Girisgin, A.O., Muz, D., & Aydin, L., 2010. Molecular detection of Nosema ceranae and Nosema apis infections in Turkish apiaries with collapsed colonies. J. Apic. Res. 49: 342. DOI: 10.3896/IBRA.1.49.4.09 [ Links ]
Office International Des Epizooties (OIE). 2008. Nosemosis of honey bees. [WWW document].URL http://www.oie.int/eng/normes/mmanual/2008/pdf/2.02.04_ NOSEMOSIS.pdf [ Links ]
Özüicli, M. & Aydin, L., 2018. Türkiye bal arilarinda ciddi tehlike; Nosemosis. Uludag Üniversitesi Veteriner Fakültesi, 37(2), 151-157. https://doi.org/10.30782/uluvfd.419001 [ Links ]
Papp, M., Békési, L., Farkas, R., Makrai, L., Maróti, G., Tozsér, D. & Solymosi, N., 2021. Natural diversity of honey bee (Apis mellifera) gut bacteriome in various climatic and seasonal states. bioRxiv. doi: https://doi.org/10.1101/2021.01.27.428438 [ Links ]
Ptaszynska, A.A., Gancarz, M., Hurd, P.J., Borsuk, G., Wiacek, D., Nawrocka, A. & Paleolog, J., 2018. Changes in the bioelement content of summer and winter western honeybees (Apis mellifera) induced by Nosema ceranae infection. PloS one, 13(7), e0200410. https://doi.org/10.1371/journal.pone.0200410 [ Links ]
Rice, R.N., 2001. Nosema disease in honeybees genetic variation and control. Rural Industries Research and Development Corporation, RIRDC Publication, UK. [ Links ]
Ricigliano, V.A., Williams, S.T. & Oliver, R., 2022. Effects of different artificial diets on commercial honey bee colony performance, health biomarkers, and gut microbiota. BMC Vet Res. 18(1), 1-14. [ Links ]
Rubanov, A., Russell, K.A., Rothman, J.A., Nieh, J.C. & McFrederick, Q.S., 2019. Intensity of Nosema ceranae infection is associated with specific honey bee gut bacteria and weakly associated with gut microbiome structure. Sci Rep. 9(1), 1-8. https://doi.org/10.1038/s41598-019-40347-6 [ Links ]
Saribiyik, C. & Özkirim, A. 2018. The transition ratio of Nosema spp. spores from colonies to honeyversus honey to colonies. J Agric Sci. 77(1), 72-80. doi:10.5539/jas.v11n1p72 [ Links ]
Sokôt, R. & Michalczyk, M., 2016. Detection of Nosema spp. in worker bees, pollen and bee bread during the honey flow season. Acta Veterinaria Brno, 85(3), 261-266. doi:10.2754/avb201685030261 [ Links ]
Somerville, D. 2005. Nosema disease in bees. Agnote DAI/124. NSW Agriculture. [ Links ]
Traver, B.E., Williams, M.R. & Fell, R.D., 2012. Comparison of within hive sampling and seasonal activity of Nosema ceranae in honey bee colonies. J. Invertebr. Pathol. 109, 187-193. https://doi.org/10.1016/j.jip.2011.11.001 [ Links ]
Tosun, O. & Yaman, M., 2016. The effects of temperature and humidity around the beehives on the distribution of Nosema ceranae, and also geographical and seasonal activity of the infection in the eastern Black Sea Region of Turkey. J. Environ. Eng. Sci B. 5, 513-522, doi:10.17265/2162-5263/2016.11.001 [ Links ]
Tunca, R.Í., Oskay, D., Gösterit, A. & Tekin., K., 2016. Does Nosema ceranae wipe out Nosema apis in Turkey? (Short Communication). Iran. J. Parasitol. 11(2):259-264. [ Links ]
Ütük, A.E., Piskin, F.Ç. & Kurt, M., 2010. Türkiye'de Nosema ceranae' nin ilk moleküler tanisi. Ankara Üniversitesi Veterinerlik Fakültesi Dergisi, 57: 275-278. [ Links ]
Wang, H., Liu, C., Liu, Z., Wang, Y., Ma, L. & Xu, B., 2020. The different dietary sugars modulate the composition of the gut microbiota in honeybee during overwintering. BMC Microbiol. 20:61 https://doi.org/10.1186/s12866-020-01726-6 [ Links ]
Whitaker, J., Szalanski, A.L. & Kence, M. 2010. Molecular detection of Nosema ceranae and N. apis from Turkish honey bees. Apidologie. doi:10.1051/apido/2010045. [ Links ]
Yarilgaç, E.S., 2016. Ordu yöresi bal arilarinin (Apis Mellifera L.) bakteriyal florasi (Master's thesis, Ordu Üniversitesi Fen Bilimleri Enstitüsü). 93 sayfa. [ Links ]
Zheng, H., Steele, M.I., Leonard, S.P., Motta, E.V. & Moran, N.A., 2018. Honey bees as models for gut microbiota research. Lab Animal, 47(11), 317. https://doi.org/10.1038/s41684-018-0173-x [ Links ]
Submitted 16 May 2022
Accepted 30 June 2022
Published 28 January 2023
# Corresponding author: rivgin@gmail.com














