Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.52 n.1 Pretoria 2022
http://dx.doi.org/10.4314/sajas.v52i1.6
ARTICLES
Polymorphism of growth hormone gene and its association with body measurement traits in Boer goat does
L.T. RashijaneI; V.G. MbazimaII; T.L. TyasiI, #
ISchool of Agricultural and Environmental Sciences, Department of Agricultural Economics and Animal Production, University of Limpopo, Private Bag X1106, Sovenga 0727, Limpopo, South Africa
IISchool of Molecular and Life Sciences, Department of Biochemistry Microbiology and Biotechnology, Private Bag X1106, Sovenga 0727, Limpopo, South Africa
ABSTRACT
The study examined single nucleotide polymorphisms (SNPs) in the growth hormone gene (GH1) in Boer goat does and their relationship with body measurement traits, namely bodyweight, body length, heart girth, rump height, rump width, ear length, cannon circumference, and head width. Seventy-six Boer goat does between the ages of 2 and 4 years were used as experimental animals. Polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) and deoxyribonucleic acid (DNA) sequencing techniques were used to detect SNPs. Chi-square test was used to measure the genetic equilibrium and a general linear model was used for the marker-trait association analysis. The PCR-RFLP and DNA sequencing results discovered one SNP (G505C) in the exon 5 of the candidate gene with two genotypes observed (AA and AB). The association analysis indicated that bodyweight was associated with the genotypes (P <0.01), but not with any of the morphometric traits. A chi-square analysis indicated that the genotypic frequencies were in Hardy-Weinberg equilibrium. The polymorphism discovered in this study is a putative marker that might assist farmers in improving their does' bodyweight through marker-assisted selection.
Keywords: bodyweight, DNA sequencing, genotype, PCR-RFLP, single nucleotide polymorphisms
Introduction
Boer goats were developed in the Eastern Cape, South Africa, in the 1900s, when a few farmers started to select crossbred and indigenous goats to produce meat (Snyman, 2014). These goats were characterized as having horns, a white solid striped forehead, long droopy ears and a brown-red colour on the head (Visser & Van Marle-Köster, 2018). They were early maturing with excellent body conformation, good carcass yield and quality, fast growth rate, and high dressing percentage and muscle to bone ratio (Pophiwa et al., 2017). They were also very fertile. At four to six months old the bucks weighed between 110 and 135 kg, whereas the does weighed between 90 and 100 kg (Pieters, 2007). McKenzie-Jakes (2007) stated that under good feeding conditions, Boer goats tended to grow by about 0.4 kg per day.
Growth hormone is an anabolic hormone that is synthesized by and released from the anterior lobe of the pituitary gland. Growth hormone regulates bone formation, muscle development and body composition in goats (Singh et al., 2015). It also has important roles in lactation, metabolism and reproduction (Alakilli et al., 2012; Malewa et al., 2014). According to Supakorn (2009), the caprine growth hormone gene (GH1) is encoded by 2.5 kbp located on the short arm of chromosome 19 (Capra hircus 19q22). It is comprised of four introns and five exons (Missohou et al., 2006). Polymorphisms in GH1 have been widely studied in some mammalian species, including Barki, Damascus, and Zaraibi goats (Boutinaud et al., 2003) Brahman cattle (Beauchemin et al., 2006), and Santa Ines (Meira et al., 2019) and Sarda (Dettori et al., 2015) sheep. However, there seems to be limited information on polymorphisms in GH1 in Boer goat does and their association with body measurement traits. However, there have been studies in various breeds of goat to investigate the effects of SNPs in GH1 (Gooki et al., 2018).
The goal of this study was to identify genetic markers in GH1 that might be useful in selection to increase the size Boer goats. Hence, the objectives were i) to identify SNPs within GH1 in Boer goat does, and ii) to assess the association of these SNPs with the weight and body measurements of Boer goat does.
Materials and Methods
The study was conducted at Tivolie Farm in Alldays, Blouberg local municipality, Limpopo, South Africa. A total of 76 Boer goat does between the ages of 2 and 4 years were used. All procedures were performed according to the standards and protocols set by the University of Limpopo Animal Research Ethics Committee (AREC) number AREC/11/2020: PG.
Blood samples were taken from the jugular vein of 76 healthy Boer goat does with the help of a veterinarian. The blood samples were collected in 10 ml ethylene diamine tetra acetic acid (EDTA) tubes and chilled until the genomic DNA was extracted. Genomic DNA was extracted and purified from the whole blood samples using a genomic DNA isolation kit (Neogen, Lansing, Michigan, USA) with minor modifications to the protocol.
A polymerase chain reaction (PCR) was performed to amplify the growth hormone gene. All primers were designed using Primer Premier 5 software (PREMIER Biosoft International, San Francisco, California, USA) and the sequence of GH1. The sequences of the forward and reverse primers for the amplification of the GH1 in the current study were as follows:
Forward: 5' - ACACCCAGGTTGCCTTCTGC -3' and
Reverse: 5' - GTCCGAGGTGCCAAACACCA -3'.
PCR was carried out in a 50 μΙ tubes containing 3 μΙ genomic DNA, 25 μΙ master mix, 1 μΙ forward, 1 μΙ reverse primers and 20 μΙ de-ionised double-distilled water. Thermal cycling conditions included an initial denaturing step of 94 °C for 5 min, followed by 32 cycles of 95 °C for 30 seconds, 52.9 °C for 30 seconds, 72 °C for 1 minute and final elongation at 72 °C for 5 minutes. The amount of 1.2% agarose gel electrophoresis was used to analyse amplicons. The gel was stained with ethidium bromide visualized and photographed under a UV trans-illuminator.
The RFLP procedure used the HaelII enzyme. It was carried out in 55 μΙ tubes containing 2 μΙ of the PCR product, 5 μΙ of 10 x buffer, 47 μΙ water and 1 μΙ of fast restriction enzyme. The reaction mixture was incubated at 36.2 °C for 1 hour. The digested products were initially separated by electrophoresis on a 1.2% agarose gel. The gel was stained with ethidium bromide and the bands were visualized and photographed under a UV trans-illuminator.
The PCR products were purified and sent to Inqaba Biotechnology Company in Pretoria, South Africa, for DNA sequencing. To identify the SNPs, sequence alignment was performed using NCBI/BLAST/blastn suite.
PCR-RFLP was performed to determine SNPs. The POPGENE software (version 1.32, University of Alberta, Canada) for population genetic analysis was used to calculate allele and genotype frequencies. A chi-square test was used to assess the allele frequencies for Hardy-Weinberg equilibrium. The GLM procedure of SAS (version 9.4, SAS Institute Inc., Cary, North Carolina, USA) was used for the marker-trait association analysis as follows:
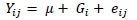
where: Yij= phenotypic values of traits (bodyweight and measurement traits),
μ = population mean,
Gi =fixed effect of genotype, and
eij = random residual error.
Results and Discussion
Descriptive statistics that characterize the Boer goat does are presented in Table 1. These goats were older and generally larger than those studied by Yousuf et al. (2020).
In this study, 654 bp designed primers were cloned for GH1. Figure 1 shows the differences in PCR amplicon sizes that resulted from these primers. For further confirmation, the PCR product sequence was compared with the DNA sequence. The nucleotide sequence findings confirmed that only two alleles were found for all the Boer goat does and indicated that the primer pair that was used amplified these GH1 alleles The PCR-RFLP banding patterns were identified for target sequence polymorphism. In all 76 Boer goats does, PCR-RFLP confirmed that the SNP in GH1 recognized AA and AB genotypes (Figure 2). DNA sequencing indicated this polymorphism was a G/C transition on the GH1 fragment at the coding region of exon 5, base 505 site (Figure 3).
The allelic and genotypic frequencies for GH1 are presented Table 2. Two alleles (A and B) were identified. The frequency of the B allele was lower than the A allele in GH1. The results also indicated that the AB genotype (0.20) showed a lower genotypic frequency than the AA genotype (0.78). Chi-square (X2) test indicated that the genotypic frequency was not significantly different (P >0.05) from the Hardy-Weinberg expectation. The effective number of alleles was 1.24 and the polymorphism information content was 0.18.
Marker-trait association results are shown in Table 3. The findings indicated that the AB genotype was associated significantly more with higher bodyweight than the AA genotype, despite there being no significant association with body length, heart girth, rump height, rump width, ear length, cannon circumference, and head width.
The current study aimed to identify genetic markers in GH1 that might be used for marker-assisted selection of the Boer goats. Similar studies by Gooki et al. (2018, 2019) and Amiri et al. (2018) observed Hardy-Weinberg equilibria in Raini Cashmere goats and Holstein cattle, respectively. However, An et al. (2010), Mahrous et al. (2018), and Gitanjli et al. (2020) reported contrary findings in Chinese goats, three Egyptian goat breeds (Barki, Damascus, Zariabi) and Gaddi goats, respectively. The association analysis findings of this study showed that AB genotypes were significantly heavier than the AA genotype, but without having a significant effect on body dimensions. DNA sequence revealed that an SNP G505C in exon 5 of the growth hormone gene.
Conclusion
The results suggest that the G505C transition in GH1 may be used to improve the bodyweight of the Boer goat does without affecting their morphometry. However, the sample size within each genotype was not large. Thus, this study provides a putative genetic marker for marker-assisted selection of Boer goats. More studies are needed to simultaneously consider sequence differences of this region and to better understand the effects of GH1 on bodyweight and morphometric traits.
Acknowledgments
The authors acknowledge the financial support from the National Research Foundation (NRF) grant number: 121987.
Authors' Contributions
LTR, VGM and TLT contributed to the project idea, design and execution of the study. LTR and TLT contributed to the acquisition of data. LTR and VGM contributed to laboratory analyses. LTR and TLT analysed the data. LTR drafted and wrote the manuscript. VGM and TLT reviewed the manuscript critically. All authors have read and approved the finalized manuscript.
Conflict of Interest Declaration
The authors declare that there is no conflict of interest relative to this work.
References
Alakilli, S.Y.M., Mahrous, K.F., Salem, L.M. & Ahmed, E.S., 2012. Genetic polymorphism of five genes associated with growth traits in goat. African J. Biotech. 11, 14738-14748. https://www.ajol.info/index.php/ajb/article/view/129449 [ Links ]
Amiri, S., Jemmali, B., Ferchichi, M.A., Jeljeli, H., Boulbaba, R. & Gara, A.B., 2018. Assessment of growth hormone gene polymorphism effects on reproductive traits in Holstein dairy cattle in Tunisia. Arch. Anim. Breed 61(4), 481-489. DOI: 10.5194/aab-61-481 [ Links ]
An, X.P., Hou, J.X., Wang, L.X., Li, G., Wang, J.G., Song,Y.X., Zhou, G.Q., Han, L., Ling, B.Y. & Cao, B.Y., 2010. Novel polymorphisms of goat growth hormone and effect on growth traits in Chinese goats. Meat Sci. 86, 758-763. DOI: 10.1016/j.meatsci.2010.06.018 [ Links ]
Beauchemin, V.R., Thomas, M.G., Franke, D.E. & Silver, G.A., 2006. Evaluation of DNA polymorphisms involving growth hormone relative to growth and carcass characteristics in Brahman steers. Genet. Mol. Res. 5, 438-447. [ Links ]
Boutinaud, M.C., Rousseau, D.H., Keisler, H. & Jammes, H., 2003. Growth hormone and milking frequency act differently on goat mammary gland in late lactation. J. Dairy Sci. 86, 509-520. DOI: 10.3168/jds.S0022-0302(03)73629-7 [ Links ]
Dettori, M.L., Pazzola, M., Pira, E., Paschino, P. & Vacca, G.M., 2015. The sheep growth hormone gene polymorphism and its effects on milk traits. J. Dairy Res. 82(2),169-176.doi: 10.1017/S0022029915000047 [ Links ]
Gitanjli, G., Sankhyan, V., Thakur, Y.P. & Dogra, P.K., 2020. Effect of growth hormone gene polymorphism on growth traits in migratory Gaddi goats of Western Himalayas, India. Tropical Anim. Health Prod. 52, 2051-2099. DOI: 10.1007/s11250-020-02227-4 [ Links ]
Gooki, F.G., Mohammadabadi, M.R. & Fozi, M.A., 2018. Polymorphism of the growth hormone gene and its effect on production and reproduction traits in goat. Iranian J. Appl. Anim. Sci. 8(4), 653-659. [ Links ]
Gooki, F.G., Mohammadabadi, M.R, Fozi, M.A. & Soflaei, M., 2019. Association of biometric traits with growth hormone gene diversity in Raini cashmere goats. Walailak J. Sci. Techn. 16(7), 499-508. [ Links ]
Mahrous, K.F., Abdel-Aziem, S.H., Abdel-Hafez, M.A.M., Abdel-Mordy, M. & Rushdi. H.E., 2019. Polymorphism of growth hormone gene in three goat breeds in Egypt. Bulletin National Res. Centre. 42(37), 2-7. DOI: 10.1186/s42269-018-0035-0 [ Links ]
Malewa, A.D., Hakin, L., Maylinda, S. & Hasain, M.H., 2014. Growth hormone gene polymorphism of Indonesia fat tailed sheep using PCR-RFLP and their relationship with growth traits. Lives. Res. Rural Dev. 26(6), 115. [ Links ]
McKenzie-Jakes, A., 2007. Selecting and evaluating goats for meat production. Getting started in the meat goat business. Bulletin 1.3. [ Links ]
Meira, A.N., Montenegro, H., Coutinho, L.L., Mouräo, G.B., Azevedo, H.C., Muniz, E.N., Machado, A.L., Sousa-Jr, L.P., Pedrosa, V.B. & Pinto, L.F.B., 2019. Single nucleotide polymorphisms in the growth hormone and IGF type-1 (IGF1) genes associated with carcass traits in Santa Ines sheep. Animal. 2019 Mar;13(3):460-468. doi: 10.1017/S1751731118001362 [ Links ]
Missohou, A., Talaki, E. & Laminon, I.M., 2006. Diversity and genetic relationships among seven West African goat breeds. Asian-Australasian J. Anim. Sci. 19, 1245-1251. https://www.animbiosci.org/journal/view.php?number=21389 [ Links ]
Pieters, A., 2007. Genetic characterization of commercial goat populations in South Africa. MSc. Dissertation, Department of Animal Breeding and Genetics, University of Pretoria, Pretoria, South Africa. [ Links ]
Pophiwa, P., Webb, E.C. & Frylinck, L., 2017. Carcass and meat quality of Boer and indigenous goats of South Africa under delayed chilling conditions. South African J. Anim. Sci. 47 (6), 794-803. c [ Links ]
Schoeman, S.J. Cloete, S.W.P. & Olivier, J.J., 2010. Returns on investment in sheep and goat breeding in South Africa. Lives. Sci. 130, 70-8. DOI: 10.1016/j.livsci.2010.02.012 [ Links ]
Singh, P.P., Tomar, S.S., Thakur, M.S. & Kumar, A., 2015. Polymorphism and association of growth hormone gene with growth traits in Sirohi and Barbari breeds of goat. Veterinary World, 8(3), 382-387. https://doi.org/10.14202/vetworld.2015.382-387 [ Links ]
Snyman, M.A., 2014. South African goat breeds: Boer goat. Info pack ref. 2014/002. Grootfontein Agricultural Development Institute. [ Links ]
Supakorn, C., 2009. The important candidate genes in goats - A review. Walailak J. Sci. Techn. 6, 17-36. DOI: 10.2004/wjst.v6i1.70 [ Links ]
Visser, C. & van Marle-Köster, E., 2018. The development and genetic improvement of South African goats. Goat Sci. 19-26. DOI: 10.5772/intechopen.70065 [ Links ]
Yousuf, F.E., Apu, A.S., Talukder, K.U., Ali, M.Y. & Husain, S.S., 2020. Adaptation and morphometric characterization of Boer goat in Bangladesh. J. Bangladesh Agric. Univ. 18(2), 428-434. https://doi.org/10.5455/JBAU98621 [ Links ]
Submitted 30 June 2021
Accepted 4 November 2021
Published 30 January 2022
# Corresponding author: louis.tyasi@ul.ac.za














