Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.50 n.4 Pretoria 2020
http://dx.doi.org/10.4314/sajas.v50i4.3
ARTICLES
Joint genetic analysis for dairy cattle performance across countries in sub-Saharan Africa
O. OpoolaI, II, III, IV, #; G. BanosI, II; J.M.K. OjangoV; R. MrodeI, V; G. SimmIII; C.B. BangaVI; L.M. BeffaVII; M.G.G. ChagundaVIII
IScotland's Rural College (SRUC), Edinburgh, United Kingdom
IIThe Roslin Institute, University of Edinburgh, United Kingdom
IIIGlobal Academy of Agriculture and Food Security, University of Edinburgh, United Kingdom
IVCentre for Tropical Livestock Genetics and Health (cTlGH), University of Edinburgh, United Kingdom
VInternational Livestock Research Institute (ILRI), Nairobi, Kenya
VIAgricultural Research Council (ARC), Pretoria, South Africa
VIILivestock Identification Trust (LIT), Harare, Zimbabwe
VIIIAnimal Breeding and Husbandry in the Tropics and Subtropics, University of Hohenheim, Stuttgart, Germany
ABSTRACT
This study assessed the feasibility of across-country genetic evaluation of dairy cattle in sub-Saharan Africa where data on livestock production are scarce. Genetic parameters were estimated for the 305-day milk yield in the first lactation and across five lactations, for age at first calving and for interval between first and second calving. Estimated breeding values of individual animals for these traits were calculated. There were records from 2 333, 25 208, and 5 929 Holstein cows in Kenya, South Africa, and Zimbabwe, and 898 and 65134 Jersey cows from Kenya and South Africa. Genetic gain from sire selection within and across countries. was predicted Genetic links between countries were determined from sires with daughters that had records in two or more countries, and from common ancestral sires across seven generations on both the maternal and paternal sides of the pedigree. Each country was treated as a trait in the across-country evaluation. The results showed that genetic variance and heritability were not always estimable within country, but were significantly different from zero in the across-country evaluation. In all three countries, there was greater genetic gain in all traits from an across-country genetic evaluation owing to greater accuracy of selection compared with within country. Kenya stood to benefit most from an across-country evaluation, followed by Zimbabwe, then South Africa. An across-country breeding programme using joint genetic evaluation would be feasible, provided that there were genetic links across countries, and would provide a platform for accelerated genetic progress through selection and germplasm exchange between sub-Saharan African countries.
Keywords: across-country genetic evaluation, genetic connectedness, genetic progress
Introduction
Genetic improvement of dairy cattle in sub-Saharan Africa has been driven mainly by the importation of foreign (exotic) breeds for pure breeding and crossbreeding with indigenous breeds (Chagunda et al., 2015). Breeds that are used for dairy production are sourced as live animals or as semen, mainly from USA, Canada, Europe, Australia and New Zealand (Maiwashe et al., 2006). Holstein is the most common exotic breed for milk production in sub-Saharan Africa (Banga, 2009), followed by Jersey, Ayrshire and Guernsey. Jerseys are popular because of the high butterfat content of their milk and their lower maintenance costs because of their smaller size. Currently, most countries of sub-Saharan Africa do not have clear breeding programmes to support dairy production owing to the lack of national livestock performance monitoring databases (Rege et al., 2011; Ojango et al., 2017). In some cases, data may exist but not be well documented. The genetic potential of dairy cattle is not fully achieved as a result of inadequate genetic improvement strategies and the lack of dynamic management systems and practices. Several studies reported low phenotypic yields and near zero genetic progress over time (Makgahlela et al., 2007; Scholtz et al., 2013).
South Africa and Kenya rank among the top milk producing countries in Africa, whereas Zimbabwe ranks in the middle (Dolecheck & Bewley, 2015). Although these countries are dispersed geographically, they are unique in their efforts to implement dairy production and to support informed breeding programmes. South Africa has the highest milk yield per cow, with opportunities to export to other countries (Chari & Ngcamu, 2019). However, there is limited government support for milk trade with other countries. South Africa is often used as the main reference point for dairy improvement in Africa because countries such as Kenya, Zimbabwe and Rwanda source South African animals to improve their national dairy herds. Kenya and Zimbabwe may also serve as reference points for genetic evaluations in other countries of Africa.
Milk production in Kenya and Zimbabwe varies through the year being generally driven by the rainfall pattern. The seasonality in the milk supply results in these countries importing milk products to meet market demands (Njarui et al., 2010; Mugweni & Muponda, 2012). Dairy productivity throughout Africa must be expanded to avoid dependence on imported dairy products. For the past three decades, South Africa has been the major driver for dairy genetic improvement strategies in Africa (Chari & Ngcamu, 2019) as a result of effective animal data and performance recording, well-organized breeding schemes, and breed-specific sire selection, which have led to genetic progress and good herd health and management. Large-scale commercial dairy farms in South Africa would be comparable with modern farms in Europe and the USA.
In Kenya, dairy production has improved through the efforts of national institutions that work in collaboration with development partners. Notable interventions from 2000 include the Smallholder Dairy Development Programme, which was implemented by the Ministry of Livestock Development in collaboration with SNV (The Netherlands), International Livestock Research Institute (ILRI), which is co-hosted by Kenya and Ethiopia, and the East Africa Dairy Development programme (EADD) (EADD, 2009; Gibson et al., 2013; SNV, 2013). On-farm production data in Kenya is maintained by the Kenya Livestock Breeders Association (KLBA), which works with national and private institutions (Kosgey et al., 2011).
Zimbabwe's dairy sector was driven by large-scale commercial producers until 2000 when land redistribution and reform programmes were implemented, leading to a drastic decline in the national dairy herd (SNV, 2016). Changing macroeconomic policies and drought-induced constraints in 2007 led to a further decline in the country's dairy sector. Smallholder dairy production in Zimbabwe is developing, although animal performance in the sector is low. There are on-going initiatives to rebuild the dairy herd with its supportive infrastructure (SNV, 2016).
The potential benefit from across-country collaboration has been well demonstrated (Banos & Smith, 1991). Opportunities to exploit genetic connectedness in across-country genetic evaluations were illustrated in analyses by Lidauer et al. (2015) of Nordic Red and Jersey, and by Pabiou et al. (2014) for Limousine and Charolais. Across-country genetic evaluations are currently available through the International Bull Genetic Evaluation Service (Interbull, Uppsala, Sweden). These evaluations are made possible through systematic genetic improvement programmes within country and collaboration between countries. South Africa is the only country from sub-Saharan Africa that currently participates in Interbull. The Interbull model, to a large extent, uses individual country genetic evaluations as the raw material for further evaluation and ranking of animals. But instead of using the Interbull model as a 'blueprint', the African scenario may well use individual performance data as the raw material for genetic evaluation, because currently few countries perform within-country genetic evaluations.
The hypothesis for the present research is that joint genetic evaluation across countries in sub-Saharan Africa will generate more accurate genetic parameters of traits and EBVs of animals, and increase the relative rate of selection response compared with national genetic evaluations. The objectives of the study were i) to calculate and compare within- and across-country genetic parameters for production and reproduction traits using data from Kenya, South Africa and Zimbabwe; ii) to estimate and compare the breeding values of individual animals from within- and across-country genetic evaluations; and iii) to quantify the predicted genetic gains from sire selection based on these genetic evaluations.
Materials and Methods
Performance and pedigree data were obtained from key livestock recording organizations in the three countries, namely the Agricultural Research Council (ARC) in Pretoria, South Africa, Kenya Livestock Breeders Association (KLBA), Kenya, and the Livestock Identification Trust (LIT) in Harare, Zimbabwe. Data comprised test-day milk production records for Holstein and Jersey cattle from 1979 to 2014. The initial data had 358 327, 10 767 516, and 260 747 test-day milk records for up to five lactations in Holstein cattle from Kenya (1979 - 2014), South Africa (1997 - 014), and Zimbabwe (1998 - 2012), respectively. Data for Jersey cattle comprised 46 242 and 1 858 021 test-day milk records for animals in Kenya and South Africa, respectively, and none for animals from Zimbabwe. The data included reproductive events for individual animals being milked, age at first calving (AFC) and the interval between first and second calvings (CI1).
The data were edited in R package (R Core Team, 2013) using these criteria. Cows whose record did not include information of the first lactation, date of birth, test-day date, date of calving, and herd location were excluded from the analysis. Age restrictions within parities were in line with those used in the South African National Dairy Genetic Evaluations (Mostert et al., 2006), whereby the maximum age at first calving is 48 months and the minimum inter-calving interval is 300 days. Lactation milk yield that was less than 305 litres was not included (Makaglela et al., 2007; Banga, 2009). Sires were required to have at least five daughters in a given country. Common sires between countries were defined as those with daughters with records in at least two of the countries. For each breed, additional genetic links were determined through common ancestors of sires that were traced over seven generations of the pedigree.
Contemporary groups were formed based on the interaction between herd, year and season of calving (HYS). In South Africa, two calendar seasons of calving were defined (Mostert et al., 2006), namely a cool dry season from April to September and a warm wet season from October to March. In Kenya, the dry season was from December to March and from July to October, whereas the wet season was from April to June and November (Ojango & Pollott, 2002). In Zimbabwe, the wet season was from September to April, whereas the dry season was from May to August (Gusha et al., 2013). An HYS group was required to have at least three cows.
Data from cows with at least six test-days (Bilal & Khan, 2009) were used to calculate the 305-day milk yield for Kenya and Zimbabwe using the test interval method (TIM) (International Committee for Animal Recording (ICAR) (ICAR, 2003). The 305-day milk records were available directly for South Africa. Hence these were not calculated.
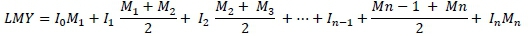
Where: = 305-day lactation milk yield (litres)
M1, M2...,Mn = kg milk yield in 24 hours on the recording day;
I1, I2...,In-1 = days between recording dates;
I0 = days between the lactation period start date and the first recording date; and
In = days between the last recording date and the 305th day of lactation.
An individual animal model was initially used to analyse 305-day milk yield in the first five lactations within country and breed using the model
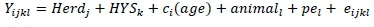
Where: Yijkl = an observation of milk produced by animal l in lactation i;
Herdj= fixed effect of herd j in which animal l was born;
HYSk= fixed effect of the kth herd-year-season in which the observation was produced;
ci(age) = fixed regression on calving age of animal l nested within lactation number ci;
animali= random additive genetic effect of animall;
pel= random permanent environment effect for animal l; and
eijkl = random error term.
The 305-day milk yield in the first lactation only was analysed with a model similar to model equation 2 without the effects of lactation number and permanent environment. Reproduction traits (AFC and CI1) were analysed with a similar model to the first lactation 305-day milk yield. The calving age was excluded from the analysis of AFC. Variance component estimates for the random effects in the model were obtained with ASReml® software (Gilmour et al., 2009) and were used to estimate the heritability and repeatability for each trait in each breed and country.
The EBVs of sires were derived through the analyses of these traits. Sire EBV reliability was computed as a function of prediction error variance for each sire and genetic variance of each trait. Subsequently, Pearson's correlations were calculated (Sedgwick, 2012) between EBVs of sires with daughters in multiple countries These correlations were adjusted for sire EBV reliability (Calo et al., 1973) to derive approximate estimates of genetic correlation between country pairs as:
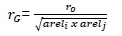
Where: rG= approximate genetic correlation;
r0= correlation between sire EBVs in the two countries;
areli= average reliability of sires in country 1; and
arelj= average reliability of sires in country 2.
Across-country analysis was implemented by pooling data from multiple countries within breed. These analyses were conducted for each trait using a model similar to equation 2 with an additional fixed effect of country. Variance components, genetic parameters and sire EBVs were derived from across-country analyses. Sire EBVs were averaged by year of birth of sire to estimate genetic trends for each trait, breed and country.
Genetic variance estimates, sire EBVs and reliabilities from the analyses were used to predict genetic gain per generation for each country, trait and breed, based on sire selection within- and across-country (Rendel & Robertson, 1950):
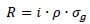
Where: R = predicted genetic gain per generation (response to selection)
i = selection intensity
p = accuracy of selection (square root of EBV reliability)
σg = square root of genetic variance estimate for a specific trait
It was assumed that sires with a minimum EBV reliability of 30% were used as selection candidates. Various selection intensities were tested based on the top 5, 10, 25, 50, 75, and 100 sires. In each case, selection intensities were calculated after accounting for the number of candidates and a finite population size. The genetic gain that was predicted to be achieved from within-country selection was compared with that from across-country selection.
Results and Discussion
After edits, data comprised 30% 75%, and 45% of the original data that had been received from Kenya, South Africa, and Zimbabwe, respectively. Seventy per cent was from the Holstein and Jersey breeds in South Africa (Table 1). The coefficient of variation for 305-day milk yield ranged from 29% to 32% in the Holstein, and was 25% in the Jersey. The average 305-day milk production by the Holsteins in South Africa was 60% higher than that in the other two countries. South African Jersey cows calved for the first time at the youngest age (861 days), and the average CI1 was lowest for animals in South Africa. The coefficient of variation for CI1 in Holsteins ranged from 22% to 42% in South Africa and Kenya. The coefficients of variation for early reproduction traits in Jerseys was lower in South Africa (AFC = 15%; CI1 = 23%) compared with Kenya (AFC = 17%; CI1 = 33%).
There were 35 and 771 Jersey sires with daughter performance records in Kenya and South Africa and 103, 505 and 236 Holstein sires with daughter performance records in Kenya, South Africa and Zimbabwe, respectively. It was evident from the pedigree data that some Holstein and Jersey sires were used across the three countries. Table 2 presents the sires and other ancestors that were common to multiple countries for Holstein and Jersey animals in terms of maternal and paternal gene flow over seven generations. Holstein had the greatest number of sires in common across countries, which were used in South Africa and Zimbabwe, followed by those in Kenya and South Africa. For the Jersey breed, one sire was common to Kenya and South Africa (Table 2), but 30 sires had common ancestry between these countries.
There were substantial additional links across the Holstein and Jersey cattle in the three countries through common ancestors. Most of the common sires of both breeds originated from USA, Canada, New Zealand, Netherlands, United Kingdom and Australia.
In some instances, it was not possible to properly de-compose the total phenotypic variance within country, which led to genetic variance and parameter estimates that were non-significantly different from zero or non-attainable (all traits in Kenya, both breeds; CI1 in Zimbabwe Holstein and AFC in South African Jersey). All the variance components were estimable in the across-country analyses and the genetic parameter estimates were significantly different from zero (P<0.05) for all traits except for AFC in the Jersey. The across-country genetic analyses yielded estimates with smaller standard errors compared with within-country. Genetic parameter estimates for milk production and reproduction traits derived within- and across-countries are presented in Table 3.
The EBVs for sires from the within-country evaluation were highly variable. The genetic trends in milk production and reproduction traits were based on the average EBV for sires from the across-country evaluation for Holsteins by country over a 15-year period (1985-2000) Although the national average milk production for Holsteins over the first five lactations within country was different (Table 1), the genetic trends for this trait in Kenya and South Africa were similar. Generally, the genetic trend in milk yield up to the fifth lactation did not show any change over the 15-year period (Figure 1). Sires in South Africa and Kenya exhibited a desirable trend in AFC (-14.21±0.51days/year) compared with Zimbabwe (Figure 2). However, there were greater fluctuations in this trait in Kenya compared with South Africa. As shown in Figure 3, undesirable positive trends were seen in Zimbabwe for CI1 (8.55 ± 1.05 days) compared with South Africa (1.84 ± 0.31 days).
The genetic trends for Jerseys for production and early reproduction traits were similar to those seen in the Holsteins for Kenya and South Africa.
Pearson's correlation between EBVs of Holstein sires from within- and across-country analyses are presented in Table 4. Correlations were not derived in cases in which the within-country genetic variance and heritability estimates were non-significant (see Table 3).
The selection intensity varied depending on the total number of sires available for selection in a country. Kenya had the lowest number for the two breeds (103 for Holsteins and 35 Jerseys), and hence had the lowest selection intensity. South Africa and Zimbabwe had 505 and 236 Holstein sires, and there were 771 Jersey sires in South Africa. However, when information on sires with common ancestry was used across the countries, the pool of candidate sires for selection increased (844 Holsteins and 806 Jerseys), enabling a higher selection intensity to be applied. The intensity of selection of the top 5, 10, 25, 50, 75 and 100 Holstein and Jersey sires that had a minimum EBV reliability of 0.30 within- and across-country is presented in Tables 5 and 6.
A standard generation interval for sires across countries was assumed. Thus predicted genetic gains for 305-day milk yield and reproduction traits from the top 5 and 10 Holstein and Jersey sires are presented in Table 7 and Table 8. No genetic gains could be predicted from within-country analysis in countries where trait genetic variance and heritability estimates were zero (all traits in Kenya, and CI1 in Zimbabwe) (Table 3). In these cases, 100% benefit would be realized from across-country genetic evaluations compared with within-country.
Phenotypically, the Holstein in South Africa and Zimbabwe had milk yield and age at calving more similar to those published by Theron and Mostert (2009) for Holsteins in South Africa. A higher age at first calving and higher milk yield than those obtained in this study were reported for Holsteins in Kenya by Muasya et al. (2014) and Menjo et al. (2009), who recorded cows calving for the first time at 1 058 days. The variations in AFC estimates may be owing to differences in the populations that were sampled. The significantly higher production of the Holsteins in South Africa relative to the other countries reflects their ability to produce high quantities of milk in sub-Saharan Africa when management conditions are probably close to optimal. The difference in milk production by Jersey cattle in Kenya and those of South Africa was not as large as that for Holsteins. Jersey cattle are smaller framed and are noted to be more adaptable to variable feeding regimes than are Holsteins. The reproductive performance of both Holstein and Jersey cattle was better in South Africa than in Kenya or Zimbabwe. There is a paucity of information on genetic evaluations of the Jersey breed in Africa.
Within-country heritability estimates for milk production in South Africa were comparable with those obtained previously for the country's Holstein population (Theron & Mostert, 2009). The estimates for first lactation milk yield were similar to those reported by Makgahlela et al. (2008). In Kenya, different heritability estimates were reported, depending on the type of production system in which the animals were reared. Ojango & Pollott (2002) reported an estimate of heritability for milk yield of 0.29 for animals raised on large-scale commercial farms, whereas Muasya et al. (2014) reported a moderate heritability of 0.17 ± 0.29 for select populations of Holsteins from medium and large-scale farms. Wasike et al. (2014) reported a heritability of 0.13 ± 0.05 and a repeatability estimate of 0.16 (SE = 0.04) for milk yield in three parities for Holstein cattle from a combination of small, medium and large-scale herds in Kenya. In Zimbabwe, heritability estimates of 0.09 (SE = 0.03) and repeatability estimates of 0.17 (SE = 0.03) over three lactations of milk yield, and a heritability estimate of 0.10 (SE = 0.09) for the first lactation milk yield in Holsteins were reported (Imbayarwo-Chikosi et al., 2001). A heritability for 305-day milk yield (0.30 SE = 0.10) was reported for Jersey cows in Zimbabwe (Missanjo, 2010), whereas in Kenya a moderate heritability estimate for milk yield in Jersey cattle of 20% was recorded (Musani & Mayer, 1997) The lack of estimable heritability for the milk yield of Jerseys in Kenya in the current study may have been because of the reduced number of records that were analysed.
Within-country heritability estimates for CI1 and AFC in South African Holstein were similar to those of a previous study on South African Holsteins (Makgahlela et al., 2007), despite differences in data. A low heritability estimate for CI1 was noted in studies on Holstein cattle raised under pasture-based systems (Olori et al., 2002). The moderate heritability for AFC in South Africa and Zimbabwe indicated potential for improvement of this trait through selection. Mostert et al. (2006) reported similar estimates for Jersey cows performing in South Africa (0.18; SE = 0.01).
Despite the limited number of direct links across the three countries in the present study, substantial links were available through common ancestors. This reflects the trade and importation of sires for genetic improvement from several regions around the world into these countries. The most common sires in the two breeds were from the USA, Canada, New Zealand, The Netherlands, UK, and Australia.
Variance components and genetic parameter estimates had higher accuracies in the across-country evaluations than within-country (Tables 7 and 8). Joint genetic evaluations increased the magnitude of the genetic parameter estimates and their accuracy, as reflected in lower standard errors associated with the estimates. Such evaluations generally help to increase the accuracy of selection where there is insufficient data in individual countries for a robust analysis. For instance, in the genetic links across these populations, the selection of sires on fertility traits for Kenya would be enhanced by joint evaluations using the larger quantity of data from South Africa. This would aid selection to attain first calving at a lower age and reduce days to second calving.
More favourable genetic trends for milk production and reproduction traits as obtained in South Africa indicated that better genetic improvement measures and breeding policies were in place compared with the other two countries. This could be a reflection of South Africa's involvement in the routine across-country gentic evaluations conducted by Interbull, combined with enhanced genetic evaluation procedures in the national dairy herd. Further improvement in the genetic trends observed in the present study requires adequate data collation and recording to enhance the effectiveness of genetic evaluations. The current results imply that slow and inconsistent genetic improvement strategies were in place, especially in Kenya and Zimbabwe. To enhance the genetic progress in various traits, farmers need to identify animals, and monitor and record their performance to inform selection decisions and advance their dairy herds positively.
Across-country genetic evaluations are routinely implemented in developed countries, based on multi-trait models with data from various populations that are treated as separate, but correlated traits (Mark et al., 2005). This accounts for the presence of genotype-by-environment interaction, which is manifested in less than unity genetic correlations between country pairs (Falconer, 1952). Although the types and amount of data to estimate genetic correlations accurately were not available, the authors derived genetic correlations based on the widely accepted method of Calo et al. (1973). These ranged from 0.58 to 0.83 for dairy sires with daughters between two countries. Mulder et al. (2006) suggested that a genetic correlation lower than 0.60 may indicate the need to develop independent breeding programmes in each environment and country. Pearson's correlations for milk production traits and the first calving interval in the current study were higher than 0.60 (Table 4), indicating that the countries would benefit from joint genetic evaluations and a common breeding programme.
Joint genetic evaluations followed by across-country selection of the best animals might have several benefits, including sharing of resources and increased genetic gains. Across-country genetic evaluations of dairy cattle may provide the tools for more effective implementation of within-country breeding programmes, and trading of genetic material. In the present study, only sires were considered, and therefore the relative rate of genetic progress represents about 66% of the possible total, and probably the most accurate, because cow information was not incorporated. Hammami et al. (2009) and Nilforooshan (2011) reported increased genetic gains from across-country genetic evaluations of performance and fertility for traits with low heritability under different environments in developed countries. In the current study, genetic progress from across-country evaluation for Jerseys was not significant (Table 8), possibly because of the small sample size from Kenya. Because of the growing dairy industry in Kenya, and the revival of the sector in Zimbabwe, these two countries would benefit most from targeted regular across-country genetic evaluation of their dairy herds. South Africa, with the largest cattle population of the three countries, could accrue benefits from cross-country evaluations through increased trade in breeding animals. These across-country genetic evaluations are the first of their kind in sub-Saharan Africa and show the potential of this approach for improving dairy production in the region.
Conclusions
The feasibility of an across-country genetic evaluation using pooled data was demonstrated. The genetic parameter estimates from across-country analyses were more accurate than within-country estimates. Countries with limited yet genetically linked within-country data would benefit most from across-country evaluations of production and fitness traits. Across-country genetic evaluations could provide robust genetic information that could enhance genetic progress and optimize future breeding strategies in sub-Saharan Africa. As more data accumulates, the best approach would be to adapt more appropriate models that reflect the diverse production environments across sub-Saharan Africa with records from countries being treated as separate but correlated traits to account for genotype by environment interaction effectively.
Acknowledgements
Data for the present study were made available from KLBA through International Livestock Research Institute (ILRI) in Kenya, Agricultural Research Council (ARC) in South Africa and Livestock Identification Trust (LIT) in Zimbabwe. The authors thank the Scottish Government for funding OO's PhD scholarship, and also their collaborators, namely JO (ILRI, Kenya), CB (ARC, South Africa) and MB (LIT, Zimbabwe), who provided access to the pedigree and performance data. MGGC, JO and RM conducted this study under the support of the African Dairy Genetic Gains (ADGG) programme, which was supported financially by the Bill & Melinda Gates Foundation. The authors thank Appolinaire Djikeng for proofreading the manuscript. The work was funded by a PhD scholarship to OO that was offered by the Scottish Government through the International Strategic Fund of SRUC (Grant number: SRUC/1026692/Chagunda).
Authors' Contributions
OO conducted the research during her PhD studies and prepared the initial draft of the manuscript. MGGC, JO, RM and GB conceived the study idea and contributed to the preparation and editing of the manuscript. MB, JO and CB also sourced and provided data from the three countries.
Conflict of Interest Declaration
The authors of this paper declare that there is no conflict of interest.
References
Banga, C.B., 2009. The development of breeding objectives for Holstein and Jersey cattle in South Africa. PhD thesis, University of the Free State, Bloemfontein, South Africa. http://scholar.ufs.ac.za:8080/xmlui/bitstream/handle/11660/648/BangaCB.pdf?sequence=1 [ Links ]
Banos, G. & Smith, C., 1991. Selecting bulls across countries to maximize genetic improvement in dairy cattle. J. Anim. Breed. Gen. 108,174-181. doi.org/10.1111/j.1439-0388.1991.tb00172.x [ Links ]
Bilal, G. & Khan, M.S., 2009. Use of test-day milk yield for genetic evaluation in dairy cattle: A review. Pakistan Veterinary Journal 29, 35-41. http://www.pvj.com.pk/pdf-files/29_1/35-41.pdf [ Links ]
Chagunda, M.G.G., Mwangwela, A., Mumba, C., Dos Anjos, F., Kawonga, B.S., Hopkins, R. & Chiwona-Kartun, L., 2015. Assessing and managing intensification in smallholder dairy systems for food and nutrition security in sub-Saharan Africa. Regional Environmental Change 1-11. doi.org/10.1007/s10113-015-0829-7 [ Links ]
Chari, F. & Ngcamu, B.S., 2019. A synthesis of risks in dairy value chains in southern Africa: Cases of South Africa and Zimbabwe. doi.org/10.5772/intechopen.84573 [ Links ]
Calo, L.L., McDowell, R.E., VanVleck, L.D. & Miller, P.D., 1973. Genetic aspects of beef production among Holstein-Friesians pedigree selected for milk production. J. Anim. Sci. 37, 676-682. doi.org/10.2527/jas1973.373676x [ Links ]
Dolecheck, K. & Bewley, J., 2015. Dairy around the world. Department of Animal and Food Sciences, University of Kentucky. https://afs.ca.uky.edu/dairy/extension/around-the-world [ Links ]
EADD, 2009. East Africa Dairy Development News No. 3. East Africa Dairy Development Project, Nairobi, Kenya. https://hdl.handle.net/10568/2444 [ Links ]
Falconer, D.S. 1952. The problem of environment and selection. American Naturalist 86, 293-298. https://www.journals.uchicago.edu/doi/abs/10.1086/281736 [ Links ]
Gibson, J.P., Rege, E., Okeyo, A.M. & Ojango, J. 2013. Overview of the Dairy Genetics East Africa (DGEA) project. Presented at the Dairy Genetics East Africa (DGEA) Project 2013 Grand Challenges Meeting, Rio de Janeiro, Brazil, 28-30 October 2013. Armidale, Australia. https://cgspace.cgiar.org/handle/10568/34851 [ Links ]
Gilmour, A.R., Gogel, B.J. & Cullis, B.R, 2009. ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead, UK. Pp 1-372. https://asreml.kb.vsni.co.uk/wp-content/uploads/sites/3/2018/02/ASReml-3-User-Guide.pdf [ Links ]
Gusha, J., Manyuchi, C.R., Imbayarwo-Chikosi, V.E., Hamandishe, V.R., Katsande, S. & Zvinorova, P.I., 2013. Production and economic performance of F1-crossbred dairy cattle fed non-conventional protein supplements in Zimbabwe. Trop. Anim. Health. Prod. 46, 1257-1263. doi.org/10.1007/s11250-013-0481-5 [ Links ]
Hammami, H., Rekik, B., Stoll, J., Bormann, J. & Gengler, N., 2009. Sustainable dairy farming - A case study of Holsteins in a developed and emerging country. IDF World Dairy Summit. Berlin, Germany. 20-24 September 2009 https://orbi.uliege.be/bitstream/2268/25845/1/WDS2009c-6.pdf [ Links ]
International Committee for Animal Recording (ICAR), 2003. Guidelines approved by the General Assembly held in Interlaken, Switzerland, on 30 May 2002, Roma, 19-39. [ Links ]
Imbayarwo-Chikosi, E.V., Makuza, S.M., Wollny, C.A.B. & Banda, J.W., 2001. Genetic and phenotypic parameters for individual cow somatic cell counts in Zimbabwean Holstein Friesian cattle. Arch. Tierz. 44, 129-37. doi.org/10.5194/aab-44-129-2001 [ Links ]
Kosgey, I.S., Mbuku, S.M., Okeyo, A.M., Amimo, J., Philipsson, J. &. Ojango, J.M.K., 2011. Institutional and organizational frameworks for dairy and beef cattle recording in Kenya: A review and opportunities for improvement. Anim. Genet. Resour. 48, 1-11. doi.org/10.1017/S2078633610001220 [ Links ]
Lidauer, M. H., Pösö, J., Pederson, J., Lassen, J., Madsen, P., Mäntysaari, E.A., Nielsen, U., Eriksson, J.Â., Johansson, K., Pitkänen, T., Stranden, I. & Aamand, G.P., 2015. Across-country test-day model evaluations for Holstein, Nordic Red Cattle, and Jersey. J. Dairy Sci. 98, 1296-1309. doi.org/10.3168/jds.2014-8307 [ Links ]
Magothe, T.M., Ilatsia, E.D., Wasike, C.B., Migose, S.A. & Kahi, A.K., 2006. Genetic evaluation of milk yield of Bos taurus dairy breeds in Kenya. In: Proceedings of the 10th KARI Scientific Conference, Nairobi, Kenya. [ Links ]
Maiwashe, A., Nephawe, K.A, Van der Westhuizen, R.R., Mostert, B.E. & Theron, H.E., 2006. Rate of inbreeding and effective population size in four major South African dairy cattle breeds. S. Afr. J. Anim. Sci. 36, 50-59. doi.org/10.4314/sajas.v36i1.3986 [ Links ]
Makgahlela, M.L., Banga, C.B., Norris, D., Dzama, K. & Ng'ambi J.W., 2007. Genetic correlations between fertility and production traits in South African Holstein cattle. S. Afr. J. Anim. Sci. 37,180-188. http://hdl.handle.net/10019.1/9326 [ Links ]
Makgahlela, M.L., Banga, C.B., Norris, D., Dzama, K. & Ng'ambi, J.W., 2008. Genetic analysis of age at first calving and calving interval in South African Holstein cattle. Asian J. Anim. Vet. Adv. 3,197-205. doi.org/10.3923/ajava.2008.197.205 [ Links ]
Mark, T., Madsen, P., Jensen, J. & Fikse, W.F., 2005. Difficulties in estimating across-country genetic correlations for weakly linked bull populations. J. Dairy Sci. 88, 3303-3305. doi.org/10.3168/jds.S0022-0302(05)73013-7 [ Links ]
Menjo, D.K., Bebe, B.O., Okeyo, A.M. & Ojango, J.M., 2009. Analysis of early survival of Holstein-Friesian heifers of diverse sire origins on commercial dairy farms in Kenya. Trop. Anim. Health Prod. 41,171-181. doi.org/10.1007/s11250-008-9172-z [ Links ]
Missanjo, E.M., 2010. Genetic and phenotypic evaluation of Zimbabwean Jersey cattle towards the development of a selection index. Published MSc thesis, University of Zimbabwe. https://www.cabi.org/gara/FullTextPDF/2013/20133184476.pdf [ Links ]
Mostert, B.E., Theron, H.E., Kanfer, F.H.J. & Van Marle-Köster, E., 2006. Test-day models for South African dairy cattle for participation in international evaluations. S. Afr. J. Anim. Sci. 36, 58-70. https://www.sasas.co.za/wp-content/uploads/2012/09/mostert36issue1hc0.pdf [ Links ]
Muasya, T.K., Peters, K.J. & Kahi, A.K., 2014. Effect of diverse sire origins and environmental sensitivity in Holstein-Friesian cattle for milk yield and fertility traits between selection and production environments in Kenya. Livestock Sci. 162, 23-30. doi.org/10.1016/j.livsci.2014.01.017 [ Links ]
Mugweni, L. & Muponda, G., 2015. Establishing the economic viability of small-scale dairy farming in Zimbabwe. IJECM 8, 448-466. http://ijecm.co.uk/wp-content/uploads/2015/08/3827.pdf [ Links ]
Mulder, H.A., Veerkamp, R.F., Ducro, B.J., Van Arendonk, J.A.M. & Bijma, P., 2006. Optimization of dairy cattle breeding programs for different environments with genotype by environment interactions. J. Dairy Sci. 89, 1740-1752. doi.org/10.3168/jds.S0022-0302(06)72242-1 [ Links ]
Musani, S.K. & Mayer, M., 1997. Genetic and environmental trends in a large commercial Jersey herd in the central Rift Valley, Kenya. Trop. Anim. Health Prod. 29, 108-116. doi.org/10.1007/BF02632330 [ Links ]
Nilforooshan, F.A. 2011. Multiple-trait multiple country genetic evaluation of fertility traits in dairy cattle. PhD thesis. https://pub.epsilon.slu.se/8094/1/Nilforooshan_ma_110427.pdf [ Links ]
Njarui, D.M.G, Gatheru, M., Wambua, J.M., Nguluu, S.N., Mwangi, D.M. & Keya, G.A.a 2010. Challenges in milk processing and marketing among dairies in the semi-arid tropical Kenya. Livest. Res. Rural Dev. 22, Article #35. http://www.lrrd.org/lrrd22/2/njar22035.htm [ Links ]
Ojango, J.M.K. & Pollott, G.E., 2002. The relationship between Holstein bull breeding values for milk yield derived in both the UK and Kenya, Livest. Prod. Sci. 74, 1-12. doi.org/10.1016/S0301-6226(01)00282-2 [ Links ]
Ojango, J.M.K., R. Mrode, R., Okeyo, A.M., Rege, J.E.O., Chagunda, M.G.G. & Kugonza, D.R., 2017. Improving smallholder dairy farming in Africa. Burleigh Dodds Science, Cambridge, UK pp. 1-26. https://hdl.handle.net/10568/78405 [ Links ]
Olori, V.E., Meuwissen, T.H.E. & Veerkamp, R.F., 2002. Calving interval and survival breeding values as a measure of cow fertility in a pasture-based production system with seasonal calving. J. Dairy Sci. 85, 689-696. doi.org/10.3168/jds.S0022-0302(02)74125-8 [ Links ]
Pabiou, T., Nilforooshan, M., Laloë, D., Hjerpe, E. & Venot, E., 2014. Across-country genetic parameters in beef cattle for interbeef weaning weight genetic evaluation. In: Proceedings 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada. 17 to 22 August 2014. https://pdfs.semanticscholar.org/084c/30d2587a5c6819d5e40ed01dbab2ca961740.pdf [ Links ]
R Core Team, 2013. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org/. [ Links ]
Rege, J.E.O., Marshall, K., Notenbaert, A., Ojango, J.M.K. & Okeyo, A.M., 2011. Pro-poor animal improvement and breeding - What can science do? Livest. Sci. 136, 15-28. https://doi.org/10.1016/jlivsci.2010.09.003 [ Links ]
Rendel, J.M. & Robertson, A., 1950. Estimation of genetic gain in milk yield by selection in a closed herd of dairy cattle. J. Genetics 50:1-8. doi.org/10.1007/BF02986789 [ Links ]
Scholtz, M.M., Van Ryssen, J.B.J., Meissner, H.H. & Laker, M.C., 2013. A South African perspective on livestock production in relation to greenhouse gases and water usage. S. Afr. J. Anim. Sci. 43, 247-254. doi.org/10.4314/sajas.v43i3.2 [ Links ]
Sedgwick, P., 2012. Pearson's correlation co-efficient. Bio. Med. Jour, 345, e4483. https://www.bmj.com/content/345/bmj.e4483 [ Links ]
SNV, 2013. Dairy sector policy study and capacity needs assessment of stakeholder associations. Kenya/Netherlands Development Organization. SNV, Nairobi, Kenya. https://agriprofocus.com/upload/Kenya Market_led_Dairy_programme1453972958.pdf [ Links ]
SNV, 2016. Zimbabwe dairy subsector study, SNV Country Office, Harare, Zimbabwe https://snv.org/cms/sites/default/files/explore/download/rarp_2016-dairy-subsector-study.pdf [ Links ]
Theron, H.E. & Mostert, B.E., 2009. Production and breeding performance of South African dairy herds. S. Afr. J. Anim. Sci. 39(Suppl. 1), 206-210. http://www.scielo.org.za/pdf/sajas/v39n5/v39n5a49.pdf [ Links ]
Wasike, C., Kahi, A.K. & Peters, K.J., 2014. Genetic relationship between lactation curve traits in the first three parities of dairy cattle. S. Afr. J. Anim. Sci. 44, 245-253. doi.org/10.4314/sajas.v44i3.6 [ Links ]
Submitted 6 February 2020
Accepted 13 April 2020
Published online 20 August 2020
# Corresponding author: oluyinka.opoola@ctlgh.org














