Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.49 n.2 Pretoria 2019
http://dx.doi.org/10.4314/sajas.v49i2.17
ARTICLES
Evaluation of candidate gene effects and environmental factors on reproductive performance of Holstein cows
S. Ardicli; H. Samli; B. Soyudal; D. Dincel; F. Balci#
Department of Genetics, Faculty of Veterinary Medicine, Uludag University, 16059 Nilufer, Bursa, Turkey
ABSTRACT
This study investigated the impact of nine polymorphisms located in the CSN2, CSN3, CSN1S1, CSN1S2, OLR1, LALBA, STAT1, DGAT1 and LGB genes, and environmental factors including calving year, season and parity on reproductive traits. The analysis was conducted on 165 Holstein-Friesian cows. Genotypes were identified using PCR-RFLP. The data of reproductive traits for four lactations were evaluated. Statistical analysis was carried out using least squares of the GLM procedures. Results indicated that CSN2 had significant effects on days before first insemination and first insemination to pregnancy interval. The SNP at the CSN3 was significantly associated with gestation length. A novel effect of OLR1-C223A on age at first calving was observed in the present study. Moreover, DGAT1 and LGB markers were significantly associated with calving interval and days before first insemination, respectively. In addition, significant environmental effects were as follows: calving year with days before first oestrus, days open, and first insemination to pregnancy interval; season with days open, first insemination to pregnancy interval, and calving interval; parity with days before first oestrus and days open. The present results and novel associations may therefore be useful and indicative for future studies on a genetic basis of cattle reproduction traits.
Keywords: cattle, environmental effects, genetic marker, Holstein-Friesian, reproduction parameters
Introduction
The health, well-being, and reproductive status of cows, along with profitability in production, are important issues in sustainable animal breeding programmes (Berglund, 2008). Genetic evaluation in dairy cattle focuses mostly on the determination of genotypic background influencing economically important traits that could be useful in the selection process including variation of milk yield and composition. However, in many countries selection for enhanced milk performance has caused a decline of cow fertility. This situation has gradually led to increased interest associated with non-production traits, including reproductive efficiency, longevity and health (Berglund, 2008; Komisarek & Dorynek, 2009). The availability of genomic data has enabled dairy cattle breeding and selection schemes. Numerous quantitative trait loci (QTL) have been proposed as candidate genes influencing various important traits in cattle (Komisarek & Dorynek, 2009; Oikonomou et al., 2009). Bovine chromosome 6 (BTA6) seems to be among the chromosomes that harbour the largest number of QTLs for milk production in cattle (Olsen et al., 2007). More than 95% of bovine milk proteins are coded by six genes: the two main whey proteins, α-lactalbumin (α-LA) and β-lactoglobulin (β-LG) are coded by the LALBA and LGB genes, and the four caseins β-CN, κ-CN, α31-0Μ and α32^Μ are coded by the CSN2, CSN3, CSN1S1 and CSN1S2 genes, respectively (Penagaricano & Khatib, 2012). The bovine LALBA gene, mapped to chromosome 5, and the LGB gene, mapped to chromosome 11 have been reported to be candidate genes for milk production and composition (Kucerova et al., 2006; Matejicek et al., 2007; Czerniawska-Piatkowska et al., 2011; Penagaricano & Khatib, 2012). The CSN2, CSN3, CSN1S1, and CSN1S2 genes belong to the casein (CN) gene family, situated on chromosome 6, and they are highly relevant in relation to milk yield and composition traits (Ikonen et al., 2001; Kucerova et al., 2006; Ibeagha-Awemu et al., 2007). The oxidised low-density lipoprotein receptor (OLR1) gene, which encodes surface receptors of vascular endothelial cells, is involved in fatty acid transport and plays an important role in the regulation of the oxidised form of low-density lipoproteins (Komisarek & Dorynek, 2009; Schennink et al., 2009). This gene is located in the interval of 106 cM to 108 cM of bovine chromosome 5, where the functional and positional candidate genes associated with fatty acid contents in milk have been mapped (Khatib et al., 2006). The signal transducer and activator of the transcription 1 (STAT1) gene encodes the cytoplasmic transcription factor (STAT1) that acts as a regulator in cytokine signalling pathways and cellular functions involving proliferation, differentiation and apoptosis (Rychtarova et al., 2014). STAT1 is located on chromosome 2 and is associated with improved milk yield and content (Khatib et al., 2009). The diacylglycerol O-acyltransferase 1 (DGAT1) gene encodes the microsomal enzyme (DGAT1) in the triglyceride synthesis, and it has recently become a highly interesting target for evaluating the fat-related traits including milk (Banos et al., 2008; Hradecka et al., 2008) and intramuscular fat content (Thaller et al., 2003).
Besides genetic structure of cattle, farm environment and management, including farm operation, calving year, calving season, parity, service period and feeding management also have significant effects on the production parameters of cows (Meikle et al., 2004; Trakovicka et al., 2013). Environmental factors directly and indirectly influence survival and productivity of dairy cattle, and the degree of environmental impact is modified by the stage-of-life cycle and adaptations of given breeds and species. To give an example, the major link between nutrition and fertility is that cows in severe negative energy balance during early lactation have lower conception rates. Energy balance is a function of feed intake and milk output and it has been shown that cows calving with a high condition score have reduced feed intakes and greater negative energy balance. It is important to note that fertility is important regardless of lactation length, and whenever insemination is planned the cow should be capable of conception (Pryce et al., 2004).
There has been growing interest in broadening selection indices to include functional traits, such as reproduction and health, because selection for production alone causes negative effects on health and reproductive performance (Miglior et al., 2005). High-yielding cows tend to be less fertile, and this extends the length of the dry period and the calving interval, as well as the rate of involuntary disposal. Less fertile cows have decreased longevity (Gonzalez-Recio et al., 2006). Besides, milk marketing systems and price constraints, or both, together with increasing producer and consumer concerns about the observed deterioration of the health and reproduction of dairy cows may be evaluated as important reasons for the above-mentioned broadening selection indices. It is worth noting that labour costs have increased relatively more than milk price in some countries (Miglior et al., 2005). Assessment of the effects of genetic markers on reproductive performance may provide important clues to pursue a balance between production and functionality in dairy cattle. To date, considerable research has been conducted on the association of genetic markers with milk yield and composition traits. However, the information about the effects of these markers on reproductive traits is rather limited (Tsiaras et al., 2005). Increased milk production has caused a decline in conception rates (Ashwell et al., 2004) and infertility has become the primary cause of involuntary culling in dairy herds (Bascom & Young, 1998). Therefore, further genetic research investigating the health and reproduction traits is needed to achieve sustainable livestock systems. In this context, the objective of the current study was to examine the impact of CN (including CSN2, CSN3, CSN1S1, and CSN1S2), OLR1, LALBA, STAT1, DGAT1 and LGB loci on reproduction traits in Holstein cows. Reproduction traits are complex because they consist of several sub-traits, and are largely influenced by the environment (Holmberg & Andersson-Eklund, 2006). Apart from genetic associations, reproductive performance is influenced by several factors such as year, season, parity and management (Tekerli & Kocak, 2009). Improving fertility would contribute to further increased production in cattle breeding. In this context, genetic contribution of fertility is very important, but non-genetic methods must also be observed in improving cattle fertility (Lazarevic & Miscevic, 2005). Therefore, an additional aim of this study was to determine the effects of environmental factors including calving year, season, and parity.
Materials and Methods
The analysis included 165 purebred Holstein-Friesian cows (Black-and-White type) from a commercial herd, located in South Marmara region (40° 15' 09.5" N and 28° 17' 59.9" E) of Turkey. Only animals with relevant data on production and reproduction parameters were used in subsequent analyses. Total herd size was 240 cows, but records with missing sire identification, incorrect calving dates and age at calving were excluded from the analysis. Milk yield of each animal was recorded daily in milking parlours equipped with electronic devices that automatically record the quantity of milk produced by individual animals. Accordingly, 305-d milk yield was calculated based on the dataset obtained from individual daily milk yield records. Regarding four lactations, mean 305-day milk production per cow was 8,732.28 ± 97.91 kg and milk yield at peak production ranged from 17.34 to 64.81 kg, with a mean of 40.56 ± 0.43 kg. The study was approved by Uludag University Local Ethics Committee for Animal Research (approval number: 2018-01/06). All animals were born in Turkey and were recorded by the Pedigree Project of the Turkish Ministry of Food, Agriculture and Livestock, and Cattle Breeders Association. Cows were housed in free-stall barns, milked thrice daily and fed ad libitum. Corn silage, alfalfa hay, oat hay and pellet-based dairy total mixed ration were formulated to meet NRC 2001 (Nutrient requirements of dairy cattle: NRC, 2001) recommendations. The data of reproductive traits from four lactations (years from 2009 to 2013) were recorded to evaluate the effects of genotypic and environmental factors. Performance measurements included days before first oestrus (DBFO) and insemination (DBFI), days open (DO), first insemination to pregnancy interval (FIPI), gestation length (GL), number of inseminations (NI), age at first calving (AFC), calving ease (CE) and calving interval (CI). Interval traits (FIPI, GL and CI) were expressed in days. GL was measured as an interval from the last insemination to the subsequent calving (Jamrozik et al., 2005). NI was determined as the number of inseminations required for conception. AFC was the age (days) of the heifer at the time of first calving. All remaining traits were defined for both heifers and cows. CE was scored according to the following criteria: 1 = unassisted calvings; 2 = calvings requiring slight assistance; 3 = calvings needing help; and 4 = Caesareans applied. Due to the low percentage of calvings scored as 4, categories 3 and 4 were joined as one (coded as 3) to minimise the extreme category problem (Gonzalez-Recio et al., 2007). All traits recorded during or before the first calving were considered heifer traits, and they were coded as parity 1. Traits measured on first-lactation animals were coded as parity 2, and the same scheme was applied to remaining (>2) lactations as described by Jamrozik et al. (2005) and Sewalem et al. (2008).
The markers in this study were selected from different sources as follows: (a) SNPs from genes that belonged to QTL mapping studies (http://www.animalgenome.org/QTLdb/cattle.html) and SNPs in genes previously associated with milk production and composition (which are potential molecular markers for reproductive performance); (b) Databases such as dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and Ensembl (http://www.ensembl.org) were used for confirmation and information of each marker.
DNA was isolated from 4 mL blood samples obtained from the jugular vein of all 165 cows using a phenol-chloroform method as described by Green & Sambrook (2012). DNA concentrations were calculated by spectrophotometer (NanoDrop 2000c, Thermo Scientific, Wilmington, DE, USA) by taking the optical density at a wavelength of 260 nm. Description of the markers evaluated in this study is presented in Table 1. All polymerase chain reactions (PCR) were carried out in a final volume of 50 μL. The reactions consisted of: 33.5 μL of ddh2O; 5 μL of 10 x buffer; 5 μL of MgSCX, (2 mM); 1 μL of dNTPs (2.5 mM); 2.5 U Taq DNA polymerase (Biomatik, A1003-500 U, 5 υ/μL); 1 μL (0.025 μΜ) of each primer; and 3 μL (approximately 70 - 160 ng) template DNA. Amplification conditions followed other authors' protocols with some modifications summarised in Table 2. PCR amplifications were run on two thermal cyclers (Palm Cycler GC1-96, Corbett Research, Sydney, Australia and MyGenie 96 Thermal Block, Bioneer, Daejeon, South Korea). Following PCR amplification, SNPs were genotyped using Restriction Fragment Length Polymorphism (RF_P) analysis. A 15 μL aliquot of the PCR product was digested with 15 units of the corresponding restriction enzyme overnight (Table 2). Digestion products were visualized through ethidium bromide (Sigma Aldrich, Steinheim, Germany) staining after electrophoresis on a 3% agarose gel (Sigma Aldrich) and visualised under UV illumination (DNR MiniLumi Gel Documentation System, Israel).
A standard chi-squared goodness-of-fit was calculated to assess compliance with Hardy-Weinberg Expectations using POPGENE software v1.32 (Yeh et al., 2000). The allelic and genotypic frequencies were calculated as described by Falconer & Mackay (1996). For each locus, heterozygosity (He), number of effective alleles (Ne) and polymorphic information content (PIC) were evaluated as population genetic parameters of polymorphisms according to formulas stated by Nei & Roychoudhury (1974) and Botstein et al. (1980). Analysis of variance (ANOVA) was used to evaluate the potential association between phenotypic characteristics and genotype groups for the adjusted residues, and association analysis was carried out by the least-squares method as applied in a general linear model (GLM) procedure of Minitab (MINITAB®, Pennsylvania, USA, v17.1.0).
For model selection we examined possible statistical models, and the best model for each phenotypic trait was selected by evaluating the adjusted R2 to compare the explanatory power of models with different numbers of predictors. Initially, the parameters such as 305-day milk yield or a cow's age were added to the model but they were excluded because of their ineffectiveness. Moreover, the interactions between environmental factors (calving year x calving season, calving year x parity, and calving season x parity) were statistically not significant and hence were excluded from subsequent analyses. Only two genotypes for locus CSN1S2 (DD and DA) and LALBA (BA and BB) were detected in the genotyped animals; moreover, there were only four and three animals with genotypes DA and BA for CSN1S2 and LALBA markers, respectively, in the examined group of animals. Hence, these markers were excluded from the statistical model in order to avoid unreliable results or confounding the influence of genotype effects on traits of interest. Association analysis was carried out for the remaining seven genes. Thus, the effects of CSN2, CSN3, CSN1S1, OLR1, STAT1, DGAT1 and LGB genotypes on days before first oestrus, days before first insemination, first insemination to pregnancy interval, days open, and number of inseminations were tested using model [1]; each trait was analysed separately:
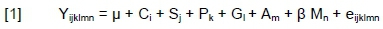
where: Yijklmn = the studied traits;
μ = the overall mean;
Ci = the fixed effect of calving year (i =1, 2, 3, 4);
Sj = the fixed effect of calving season (j = autumn, winter, spring and summer);
Pk = the fixed effect of parity (k = 1, 2, 3, 4);
Gl = fixed effect of the lth genotype for CSN2 (l = A1A1, A1A2, A2A2), CSN3 (l = AA, AB, BB), CSN1S1 (l = BB, BC), OLR1 (l = AA, AC), STAT1 (l = CC, CT, TT), DGAT1 genotype (l = KA, KK), or LGB (l = AA, AB, BB);
Am = random effect of animal factor (m = varying performance values of an individual cow in different lactations);
β Mn = the regression effect of peak milk yield on selected reproductive traits and
eijklmn= the random residual effect.
Model [2] was used to test the effects of CSN2, CSN3, CSN1S1, OLR1, STAT1, DGAT1 and LGB genotypes on age at first calving, gestation length, calving interval, and calving ease; each trait was analysed separately:
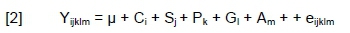
where: Yijklm = the studied traits; μ = the overall mean;
Ci = the fixed effect of calving year (i =1, 2, 3, 4);
Sj = the fixed effect of calving season (j = autumn, winter, spring and summer);
Pk = the fixed effect of parity (k = 1, 2, 3, 4);
Gl = fixed effect of the lth genotype for CSN2 (l = A1A1, A1A2, A2A2), CSN3 (l = AA, AB, BB),
CSN1S1 (l = BB, BC), OLR1 (l = AA, AC), STAT1 (l = CC, CT, TT), DGAT1 genotype (l = KA, KK), or LGB (l = AA, AB, BB);
Am = random effect of animal factor (m = varying performance values of an individual cow in different lactations); and
eijklm = the random residual effect.
For all statistical comparisons, a probability level of P <0.05 was accepted as statistically significant. In addition, the P-value less than 0.10 (P <0.1) was, therefore, considered as a tendency (a suggestive association). When significant associations were identified, the Tukey's multiple comparison test was applied to reveal differences.
Results
Two alleles and three genotypes in the CSN2, CSN3, STAT1 and LGB were found, whereas only two genotypes were present in the remaining markers. The genotype and allele frequencies as well as the population genetic parameters including He, Ne, PIC, and compatibility with the Hardy-Weinberg equilibrium (HWE) are shown in Table 3. The minor allele frequencies (MAF) ranged from 0.01 to 0.48 and markers CSN1S2 and LALBA showed extremely low MAF values (<0.01). At locus CSN1S2, genotype DD and at locus LALBA, genotype BB exhibited a frequency higher than 0.97, being nearly fixed in the Holstein population.
Deviation from HWE was tested for each locus and was not compatible for OLR1, STAT1 and DGAT1 markers (Table 3). The He values ranging from 0.0196 to 0.4992; Ne values ranging from 1.0202 to 1.9968; and PIC values ranging from 0.0194 to 0.3746 were observed in the chi-square statistics.
The general linear model (GLM) analysis revealed that all polymorphic markers tested, with the exception of STAT1, showed associations with reproduction traits, comprising six significant associations (P <0.05) and one suggestive association (P <0.1) in total (Table 4). The SNPs in the CSN1S2 and LALBA genes did not segregate in the studied population (n = 165 genotyped) and were therefore excluded from the statistical model. Table 5 shows the least-squares means and their respective standard errors. Results revealed that the CSN2 A1A1 genotype was significantly associated with the shorter DBFI and FIPI (P <0.05). The CSN3 marker affected the GL and BB genotype and was associated with the shorter GL, compared to alternative variants. A novel effect of OLR1-C223A on AFC was observed. In this context, the AA genotype was significantly associated with the higher AFC, compared to heterozygotes (Table 5). In addition, there were significant effects of DGAT1 and LGB markers on CI and DBFI, respectively. There was no association between the CSN1S1 and STAT1 markers and any of the traits evaluated, nor was there any association with the tested SNPs with DBFO, DO, NI and CE.
Calving year, calving season and parity affected the reproductive performance traits at different levels of significance, as shown in Table 4. Accordingly, least-squares means and standard errors for the impact of environmental factors on reproductive performance traits in Holstein cows raised in Turkey are presented in Table 6. Results revealed that DBFO, FIPI and DO were significantly affected by calving year. The calving season showed significant effects on FIPI and DO. Moreover, parity was significantly associated with differences in DBFO and DO. No statistically significant differences were observed in AFC, DBFI, NI, GL, and CE.
Discussion
In the current study, all loci were consistent with HWE, except deviations for OLR1, STAT1, and DGAT1 markers. Deviations from HWE may result from either the population substructure, the presence of null alleles and high selection pressure; moreover, inbreeding or indirect selection for these loci from the selection for milk production in the Holstein breed may be another explanation (Loftus et al., 1999; Lacorte et al., 2006). If diallelic loci have allele frequencies less than 0.95; these can be evaluated as polymorphic (Menezes et al., 2006). Accordingly, markers used in this study may be classified as polymorphic except for the CSN1S1, CSN1S2 and LALBA. Population genetic parameters including He, Ne, and PIC comprise the evaluation of population structure defined by genetic variation of a particular gene or genes; for instance, the low heterozygosity values indicate that inbreeding probably may be a potential problem at the population level, and therefore should be taken into account in pedigree evaluation. Besides, the effectiveness of loci allele impact in populations has been expressed by Ne (Trakovicka et al., 2013). Results indicated that comparison of He and Ne showed higher gene heterozygosity (>0.46) and effective allele numbers (>1.85) across the population for CSN2, OLR1, STAT1, DGAT1 and LGB markers, and indicated a good level of genetic variability in the analysed cow population at the considered locus. The PIC values indicate the quality of markers in genetic studies (Botstein et al., 1980). According to the classification of PIC (low polymorphism if PIC value is <0.25, median polymorphism if PIC value is between 0.25 - 0.5, and high polymorphism if PIC value is >0.5) (Botstein et al., 1980), the analyses showed low (CSN3, CSN1S1, CSN1S2 and LALBA) or median (CSN2, OLR1, STAT1, DGAT1 and LGB) levels of PIC of analysed SNPs across the present population based on the expected heterozygosity. Remarkable differences in the genotype frequency distributions and population genetic parameters exist in contemporary literature for the markers used in this study in various cattle breeds. Genotypic distribution and structure may vary among breeds and even among different populations of the same breed. Inbreeding, population stratification and artificial selection of parents for the milk production traits in different herds may also influence the population characteristics (Lacorte et al., 2006).
There are several indications, implying that CN (gene family), OLR1, STAT1, DGAT1 and LGB may be strong candidate genes controlling dairy cattle production including milk yield and components. However, studies about the associations between the mentioned genes and reproduction traits are relatively limited. Improvement without the need for DNA marker information of health and reproduction traits, which express low heritability, is rather difficult to achieve, compared to milk production in dairy cattle (Ashwell et al., 2004). Analyses of increased genetic gain using marker-assisted selection (MAS) can provide a trustworthy genetic evaluation and a shortened generation interval, required for realisation in cattle breeding (Trakovicka et al., 2013). In the present study, we intended to determine whether DNA markers commonly studied for milk production traits could be applied via reproduction traits in Holstein breed, and to evaluate the potential novel genetic-associations. Our results revealed that CSN2, CSN3, OLR1, DGAT1 and LGB genes may be associated with reproduction performance. Among them, CSN2 significantly affected the DBFI and FIPI (P <0.05). Animals with the A2A2 genotype had higher means than A1A2 and A1A1 genotypes for DBFI (+5.12 days and +11.43 days, respectively) and FIPI (+3.62 days and +4.63 days, respectively) traits. These results may indicate a potential association of CSN2 genotypes with reproduction traits.
Demeter et al. (2010) and Penagaricano & Khatib (2012) did not detect significant associations with CSN2 and cow fertility traits such as CI, rate of calving after first insemination, and number of services per conception in Holsteins. Moreover, Demeter et al. (2010) suggested that a breeding programme aiming to improve the manufacturing properties of milk by selecting animals based on milk protein variants would have no negative influence on the reproductive performance of cows. However, here we report that CSN2 genotypes influenced DBFI and FIPI. CSN2 variants play an important role in milk and cheese yield/quality and their concentration is positively associated with good rennet properties of milk (Massella et al., 2017). Moreover, A2 milk (A1 free) is produced in many different countries, claiming its potential benefits in human health (Caroli et al., 2016; Brooke-Taylor et al., 2017), but the studies about positive effects of A2 milk or unfavourable effects of A1 milk are often conflicting. Apart from the contradictory human health studies about CSN2 genotypes (A1/A2), there is still a strong need for the evaluation of corresponding genotype effects on cow fertility and reproduction performance.
The present results indicated that the CSN3 marker significantly affected the GL (P <0.05). Animals with AA genotype had +5.77 days and +5.08 days longer GL compared to BB and heterozygous animals, respectively. On the other hand, contradictory results (no significant association between the CSN3 genotypes and reproduction traits) have also been reported in previous studies performed by Tsiaras et al. (2005), Demeter et al. (2010) and Penagaricano & Khatib (2012). Some researchers also reported a tendency towards lower AFC, shorter CI and inter-pregnancy interval (Tsiaras et al., 2005; Felenczak et al., 2008a). GL is one of the decisive parameters for cattle reproduction and GLs longer or shorter than average are associated with increased problems at parturition (Silva et al., 1992). It is worth noting that the AA genotype frequency was very low; moreover, there were only seven animals carrying this genotype in the present study. Hence, further genetic studies about the association of CSN3 with GL should be conducted for more reliable conclusions.
Although there were evident differences in the AFC values of cows with different CSN1S1 genotypes, this could not be statistically substantiated. Animals with the BB genotype reached the AFC 46.80 days earlier than heterozygous animals. AFC includes the period that a cow needs to reach maturity and to reproduce for the first time (Hossein-Zadeh, 2011) and it can be evaluated as an important factor in the cost of rearing replacements in dairy herds to maximise lactation performance and reduce rearing costs (Ettema & Santos, 2004). Apart from the tendency observed for CSN1S1 genotypes, a statistically significant effect of OLR1 marker on AFC was determined in the present study. Cows with the AA genotype were characterised by a longer AFC (+49.20 days) in comparison with heterozygotes. To achieve maximum lactation and reproduction performance, average AFC in Holsteins was recommended to be <24 months with body weight >560 kg after calving at 24 months (Heinrichs, 1993; Tozer & Heinrichs, 2001). AFC in Holsteins can be categorised into three groups: low (<700 days), medium (701 - 750 days), and high (>751 days) AFC, as described by Ettema & Santos (2004). According to this classification, genotype means indicated high AFC for both AA and AC. AFC can affect milk production traits and productive life in dairy cattle (Nilforooshan & Edriss, 2004). Hence, results obtained from genetic association studies about AFC may provide novel aspects to achieve sustainable dairy management. We report in this study that OLR1-C223A may affect AFC in the Holstein breed. On the contrary, Komisarek & Dorynek (2009) and Rychtarova et al. (2014) found no significant association between this marker and any of the reproduction traits analysed The OLR1 gene influences glucose and lipid metabolisms because it encodes a vascular endothelial cell-surface receptor that binds and degrades the oxidised forms of low-density lipoproteins (Mehta & Li, 2002). Thus, the existence of a novel association between OLR1-C223A and reproduction parameters in cattle would not be surprising.
Selection for increased milk production, which is unfavourably correlated with fertility, has resulted in undesirable genetic trends in reproduction performance (Royal et al., 2002). In this context, many genes associated with milk production traits have been identified, but their effects on fertility and reproduction performance have not been evaluated equally. The DGAT1 gene appears to be a strong candidate gene for determining milk production and reproductive traits according to several studies conducted on various cattle breeds. In this study, the DGAT1-K232A marker was significantly associated with CI (P <0.05). Cows with the KK genotype were characterised by a longer CI (+70.9 days) in comparison with heterozygotes. To achieve maximum economic benefits, optimum CI should be 350 - 380 days (Kopuzlu et al., 2008); accordingly, the present results revealed an unfavourable CI for the KK genotype. In accordance with our results, Rychtarova et al. (2014) reported that DGAT1 had a significant effect on CI (P <0.05). However, these researchers found that the KK genotype was associated with better results for CI (392.7 ± 4.03 and 377.6 ± 6.91 for KA and KK genotypes, respectively) and this genotype was also effective on DO, days from first to last insemination, and NI, comparable to the results obtained by Oikonomou et al. (2009). On the other hand, Demeter et al. (2009) found a potential effect of the DGAT1 on non-return rates for insemination 28 and 56 days after the first service. Collis et al. (2012) suggested that the K232A polymorphism in the gene DGAT1 was associated with age at puberty. Conversely, Berry et al. (2010) indicated that there was no association between the K232A marker and fertility and calving performance traits in Irish Holstein-Friesians. The DGAT1 gene encodes the microsomal enzyme (DGAT1) in the triglyceride synthesis and the mechanism by which the DGAT1-K232A polymorphism influences reproduction traits may be based on the metabolic activities with regard to lipid synthesis. Nevertheless, there is still room for improvement in understanding the genetic basis of cattle reproduction.
ANOVA results showed that LGB genotypes made significant changes in DBFI (P < 0.05). The shortest DBFI was found in cows with the BB genotype and the highest in AA homozygotes, the difference being 10.08 days. Fewer studies reported the possible association of LGB polymorphisms with reproductive traits compared to milk production-related studies. Tsiaras et al. (2005) and Felenczak et al. (2008b) reported a tendency towards lower AFC in cows with the LGB BB genotype and longer CI compared to cows with the AA genotype. Jairam & Nair (1983) and Lin et al. (1987) also reported lower AFC in cows with the LGB AB genotype. Penagaricano & Khatib (2012) suggested that polymorphisms in the LGB gene showed significant associations with fertilisation success and blastocyst rate. In contrast, Ruottinen et al. (2004) and Demeter et al. (2010) found no significant relationship between LGB variants with several fertility traits in Finnish Ayrshire and Holstein dairy cattle, respectively. Although high and antagonistic genetic correlations between protein concentration or protein yield and fertility have been previously reported, knowledge of the association between milk protein variants and cow fertility is limited and in some cases contradictory (Penagaricano & Khatib, 2012). Hence, further genetic experiments may be useful to document the results providing not only milk yield/components but also non-production traits, such as cow fertility and reproduction performance to acquire more effective and sustainable selection schemes.
Reproduction traits in dairy cattle are influenced by many environmental factors. In the current study, apart from genetic marker associations, we also examined the effects of calving year, season and parity. Results revealed that the first year was associated with better results for DBFO, DO and FIPI. Days open is one of the most important indicators that display the status of reproduction management which should be 70 - 90 days at optimum for a profitable enterprise. In the present study, the fourth year was associated with an unfavourable DO. The differences observed in DO according to calving year variable could be related to the variation in management practices; the same explanations apply to DBFO and FIPI. Indeed, DBFO and FIPI are the basic constituents for the calculation of DO. In the literature, the year effect on different reproduction traits in cattle breeding has been reported. Lazarevic & Miscevic (2005) suggested that the year effect has significantly (P <0.05) expressed action on the traits of duration of pregnancy, as well as on the trait of duration of the service period. Tekerli & Kocak (2009) reported that calving year had a significant effect (P <0.05) on breeding efficiency (%), herd life and length of productive life. Conversely, Balci (1999) reported that calving year did not affect any of the reproductive traits in Holstein cows. The effect of year should be taken into consideration as a decisive parameter when evaluating the reproduction performance of cows, as the production level and management factors obviously change through years according to the breeding strategies.
In this study, the effect of season showed a high significance (P <0.001) on investigated traits of reproduction including DO, FIPI and CI. Longest intervals were consistently recognised in the summer season; moreover, spring was associated with better results for the mentioned traits. The differences between best and worst calving season accounted for 71.3 (DO), 43.1 (FIPI) and 92.4 days (CI), respectively. Bielfeldt et al. (2004), similar to our results, observed obvious tendencies for the effect of calving season on fertility traits including days to first service, DO and CI, and the longest intervals were determined in season April to June in Swiss Brown cows. However, these researchers found that January to March season was only marginally better, and that shorter intervals were observed by far in cows calving in the second half of the year. Lazarevic & Miscevic (2005) reported that season (summer) had a significant effect (only for second generation: daughters) on duration of pregnancy (expressed as GL in our study), service period (expressed as DO in our study), and interval between calvings (expressed as CI in our study) (P <0.05). These researchers also found that spring had an effect on the duration of pregnancy (only for third generation: granddaughters) (P <0.05), summer affected the duration of service period and interval between calvings (P <0.05), and winter had an effect on the duration of service period (P < 0.05). However, Ray et al. (1992) indicated that cows calving in spring and summer had reduced reproductive performance, as measured by CI and services per conception (expressed as NI in our study). Tekerli & Kocak (2009) reported a significant effect of season on breeding efficiency (higher values for autumn and winter) (P <0.05). Apart from the significant associations, we report herein a tendency for the effect of calving season on CE (P <0.1). Winter season appeared to be associated with better results for CE but the variations among calving seasons were low. Calving season is one of several constituents contributing to environmental factors. However, it is difficult to compare fertility parameters between different geographical locations due to varying seasonal factors. Besides the level of nutrition and exercise, photoperiodism and temperature may be considered as other important factors responsible for seasonal variation in fertility (Bielfeldt et al., 2004).
Our study clearly demonstrates that parity has significant effects on DBFO and DO (P <0.05). Markedly the highest values for the mentioned traits were observed in animals of the first parity. Moreover, the lowest values for DBFO and DO were associated with the third (-21.70 days) and the fourth parities (-20 days), respectively. Although shown to be statistically non-significant, first parity cows tended to have longer FIPI compared to cows with parities 2 and above (P <0.1). Goshu et al. (2007) also found that the effect of parity was highly significant (P <0.001) for DO and it affected the CI and NI as well. Results of these researchers indicated that DO and CI decreased as parity numbers increased until three and the first parity cows required 0.36 less NI than the population average. Similarly, Ray et al. (1992) suggested that first lactation cows had the lowest values for CI and NI. Balendran et al. (2008) demonstrated that parity had a significant effect on pregnancy rate (PR) and lower PR was observed in animals of higher parity, which is in agreement with related studies reported by Butler & Smith (1989) and Pryce et al. (2004). Meikle et al. (2004) reported that effect of parity was effective on the initiation of the ovarian cycle, parturition to first service, and parturition to conception intervals. They also demonstrated that parity affects non-esterified fatty acids, thyroid hormones (T3 and T4) and insulin-like growth factor-I (IGF-I) concentrations as well as reproductive parameters in dairy cows. In addition, Lee & Kim (2006) indicated that the culling rate due to reproductive failure increased with parity. The effects of parity on reproductive performance-related traits and fertility may be based on the follicular development, circulating hormone concentrations, energy balance, and postpartum reproductive function in dairy cows.
Conclusion
The results of the present study indicate that the CSN2, CSN3, OLR1, DGAT1 and LGB genotypes influence certain reproduction traits in Holstein cows. The A1A1 CSN2 genotype had positive effects on days before first insemination and first insemination to pregnancy interval. The BB variant of CSN3 was associated with shorter gestation length, and the heterozygote genotype of OLR1 was associated with lower age at first calving. The DGAT1 KA genotype and LGB B allele had positive effects on calving interval and days before first insemination, respectively. Therefore, these genotypes appear to be potential candidates for the selection of reproduction trait improvement. However, further studies performed with larger populations may be needed to confirm these associations. Reproductive performance was virtually unaffected by STAT1 genotypes. Calving year with days before first oestrus, days open, and first insemination to pregnancy interval; season with days open and first insemination to pregnancy interval and calving interval; parity with days before first oestrus and days open were determined as significant environmental effects. Results of this study suggest that a balance between production and functionality must be pursued, and proper economic weights must be applied to every trait because selection for favourable milk-related genes is expected to influence the reproductive performance of dairy cattle. Selection indices have evolved worldwide, shifting the focus from production to a more balanced breeding goal of improving production together with conformation and reproduction. This study represents important points which may provide an adequate genotypic evaluation of dairy cattle reproduction performance. The broadening of breeding goals with respect to changes in selection indices may be more useful than conventional production-focused aspects to achieve sustainable and profitable dairy cattle management.
Acknowledgements
The authors would like to thank "Matli Food Group" for their help in managing the animals and collecting blood samples. The DNA samples used in this study were obtained from a project which was supported by Scientific Research Projects Funds of Uludag University with grant no: UAPV-2011/21. The authors thank I.Taci Cangul for language assistance.
Authors' Contributions
SA and FB conducted the research and were responsible for drafting and submitting the manuscript. SA, HS, BS, DD were responsible for laboratory analyses. SA, HS, FB critically analysed interpreted results and assisted in the revision of the original and revised manuscripts. FB, SA was responsible for statistical analyses and BS, together with SA, assisted in animal husbandry.
Conflict of Interest Declaration
There are no conflicts of interest.
References
Ashwell, M., Heyen, D., Sonstegard, T., Van Tassell, C., Da, Y., VanRaden, P., Ron, M., Weller, J. & Lewin, H., 2004. Detection of quantitative trait loci affecting milk production, health, and reproductive traits in Holstein cattle. J. Dairy Sci. 87, 468-475. [ Links ]
Balci, F., 1999. The effects of year, calving number and calving season on some reproductive characteristics of Holstein cattle. Uludag Univ. J. Fac. Vet. Med. 18, 239-249 (in Turkish, English abstract). [ Links ]
Balendran, A., Gordon, M., Pretheeban, T., Singh, R., Perera, R. & Rajamahedran, R., 2008. Decreased fertility with increasing parity in lactating dairy cows. Can. J. Anim. Sci. 88, 425-428. [ Links ]
Banos, G., Woolliams, J., Woodward, B., Forbes, A. & Coffey M., 2008. Impact of single nucleotide polymorphisms in leptin, leptin receptor, growth hormone receptor, and diacylglycerol acyltransferase (DGAT1) gene loci on milk production, feed, and body energy traits of UK dairy cows. J. Dairy Sci. 91, 3190-3200. [ Links ]
Bascom, S. & Young, A., 1998. A summary of the reasons why farmers cull cows. J. Dairy Sci. 81, 2299-2305. [ Links ]
Berglund, B., 2008. Genetic improvement of dairy cow reproductive performance. Reprod. Domest. Anim. 43, 89-95. [ Links ]
Berry, D.P., Howard, D., O'Boyle, P., Waters, S., Kearney, J. & McCabe, M., 2010. Associations between the K232A polymorphism in the diacylglycerol-O-transferase 1 (DGAT1) gene and performance in Irish Holstein-Friesian dairy cattle. Irish J. Agr. Food Res. 49, 1-9. [ Links ]
Bielfeldt, J., Badertscher, R., Tólle, K.-H. & Krieter, J., 2004. Influence of systematic effects on fertility traits in Swiss Brown cows. Arch. Anim. Breed. 47, 537-549. [ Links ]
Botstein, D., White, R.L., Skolnick, M. & Davis, R.W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314-331. [ Links ]
Brooke-Taylor, S., Dwyer, K., Woodford, K. & Kost, N., 2017. Systematic Review of the Gastrointestinal Effects of A1 Compared with A2 β-Casein. Adv. Nutr. 8, 739-748. [ Links ]
Butler, W. & Smith, R., 1989. Interrelationships between energy balance and postpartum reproductive function in dairy cattle. J. Dairy Sci. 72, 767-783. [ Links ]
Caroli, A.M., Savino, S., Bulgari, O. & Monti, E., 2016. Detecting β-casein variation in bovine milk. Molecules 21, 141-147. [ Links ]
Collis, E., Fortes, M., Zhang, Y., Tier, B., Schutt, K., Barendse, W. & Hawken, R., 2012. Genetic variants affecting meat and milk production traits appear to have effects on reproduction traits in cattle. Anim. Genet. 43, 442-446. [ Links ]
Czerniawska-Piatkowska, E., Szewczuk, M., Olszewska, A. & Chocilowicz, E., 2011. Association between beta- lactoglobulin (LGB) polymorphism and yield and composition of milk Holstein-Friesian cows imported from Sweden. Acta Sci. Pol. Zootechnica 10, 9-18. [ Links ]
Demeter, R. & Markiewicz, K., Van Arendonk, J. & Bovenhuis, H., 2010. Relationships between milk protein composition, milk protein variants, and cow fertility traits in Dutch Holstein-Friesian cattle. J. Dairy Sci. 93, 5495-5502. [ Links ]
Demeter, R., Schopen, G., Lansink, A.O., Meuwissen, M. & Van Arendonk, J. 2009. Effects of milk fat composition, DGAT1, and SCD1 on fertility traits in Dutch Holstein cattle. J. Dairy Sci. 92, 5720-5729. [ Links ]
Ettema, J. & Santos, J., 2004. Impact of age at calving on lactation, reproduction, health, and income in first-parity Holsteins on commercial farms. J. Dairy Sci. 87, 2730-2742. [ Links ]
Falconer, D.S. & Mackay T.F.C., 1996. Introduction to Quantitative Genetics, 4th ed.. Harlow, England. Pearson Education Ltd. p 280. [ Links ]
Felenczak, A., Gil, Z., Adamczyk, K., Zapletal, P. & Frelich, J., 2008a. Polymorphism of milk κ-casein with regard to milk yield and reproductive traits of Simmental cows. J. Agrobiol. 25, 201-207. [ Links ]
Felenczak, A., Jezowit-Jurek, M., Gil, Z. & Adamczyk, K., 2008b. Polymorphism of milk β-lactoglobulin and its effect on milk yield and reproductive traits of Simmental cows. Ann. Anim. Sci. 8, 207-213. [ Links ]
Gonzalez-Recio, O., Alenda, R., Chang, Y.M., Weigel, K.A. & Gianola, D., 2006. Selection for female fertility using censored fertility traits and investigation of the relationship with milk production. J. Dairy Sci. 89, 4438-4444. [ Links ]
Gonzalez-Recio, O., De Maturana, E.L. & Gutierrez, J., 2007. Inbreeding depression on female fertility and calving ease in Spanish dairy cattle. J. Dairy Sci. 90, 5744-5752. [ Links ]
Goshu, G., Belihu, K. & Berihun, A., 2007. Effect of parity, season and year on reproductive performance and herd life of Friesian cows at Stella private dairy farm, Ethiopia. Livest. Res. Rural Dev., 19. Available from http://www.lrrd.org/lrrd19/7/gosh19098.htm (Accessed 07.05.2019). [ Links ]
Green, M.R. & Sambrook, J., 2012. Isolation of high-molecular-weight DNA from mammalian cells using proteinase K and phenol. In: Molecular Cloning: A Laboratory Manual, 4. Cold Spring Harbor, New York, USA. Cold Spring Harbor Laboratory Press . pp. 47-48. [ Links ]
Heinrichs, A., 1993. Raising dairy replacements to meet the needs of the 21st century. J. Dairy Sci. 76, 3179-3187. [ Links ]
Holmberg, M. & Andersson-Eklund, L., 2006. Quantitative trait loci affecting fertility and calving traits in swedish dairy cattle. J. Dairy Sci. 89, 3664-3671. [ Links ]
Hossein-Zadeh, N.G., 2011. Genetic and phenotypic trends for age at first calving and milk yield and compositions in Holstein dairy cows. Arch. Tierz. 4, 338-347. [ Links ]
Hradecka, E., Citek, J., Panicke, L., Rehout, V. & Hanusova, L., 2008. The relation of GH1, GHR and DGAT1 polymorphisms with estimated breeding values for milk production traits of German Holstein sires. Czech J. Anim. Sci. 53, 238-245. [ Links ]
Ibeagha-Awemu, E., Prinzenberg, E.-M., Jann, O., Lühken, G., Ibeagha, A., Zhao, X. & Erhardt, G., 2007. Molecular characterization of bovine CSN1S2*B and extensive distribution of zebu-specific milk protein alleles in European cattle. J. Dairy Sci. 90, 3522-3529. [ Links ]
Ikonen, T., Bovenhuis, H., Ojala, M., Ruottinen, O. & Georges, M., 2001. Associations between milk protein polymorphism and first lactation milk production traits in Finnish Ayrshire cows. J. Dairy Sci. 82, 1026-1033. [ Links ]
Jairam, B. & Nair, P., 1983. Genetic polymorphism of milk proteins and economic characters in dairy animals. Indian J. Dairy Sci. 53, 1-8. [ Links ]
Jamrozik, J., Fatehi, J., Kistemaker, G. & Schaeffer, L., 2005. Estimates of genetic parameters for Canadian Holstein female reproduction traits. J. Dairy Sci. 88, 2199-2208. [ Links ]
Khatib, H., Huang, W., Mikheil, D., Schutzkus, V. & Monson, R., 2009. Effects of signal transducer and activator of transcription (STAT) genes STAT1 and STAT3 genotypic combinations on fertilization and embryonic survival rates in Holstein cattle. J. Dairy Sci. 92, 6186-6191. [ Links ]
Khatib, H., Leonard, S., Schutzkus, V., Luo, W. & Chang, Y., 2006. Association of the OLR1 gene with milk composition in Holstein dairy cattle. J. Dairy Sci. 89, 1753-1760. [ Links ]
Komisarek, J. & Dorynek Z., 2009. Effect ofABCG2, PPARGC1A, OLR1 and SCD1 gene polymorphism on estimated breeding values for functional and production traits in Polish Holstein-Friesian bulls. J. Appl. Genet. 50, 125-132. [ Links ]
Kopuzlu, S., Emsen, H., Ozluturk, A. & Kucukdemir, A., 2008. Esmer ve Siyah Alaca irki sigirlarin Dogu Anadolu Tarimsal Araçtirma Enstitusu sartlarinda dol verim ozellikleri, Lalahan Hayvanc. Arast. Enst. Derg. 48, 13-24 (in Turkish, English abstract). [ Links ]
Kucerova, J., Matejicek, A., Jandurova, O., Sorensen, P., Nemcova, E., Stipkova, M., Kott, T., Bouska, J. & Frelich, J., 2006. Milk protein genes CSN1S1, CSN2, CSN3, LGB and their relation to genetic values of milk production parameters in Czech Fleckvieh. Czech J. Anim. Sci. 51, 241-247. [ Links ]
Lacorte, G., Machado, M., Martinez, M., Campos, A., Maciel, R., Verneque, R., Teodoro, R., Peixoto, M., Carvalho, M. & Fonseca, C., 2006. DGAT1 K232A polymorphism in Brazilian cattle breeds. Genet. Mol. Res. 5, 475-482. [ Links ]
Lazarevic, R. & Miscevic, B., 2005. Inheritance of some fertility traits in three successive generations of Holstein-Friesian cattle. Arch. Tierz. 48, 5-11. [ Links ]
Lee, J.-Y. & Kim, I.-H., 2006. Advancing parity is associated with high milk production at the cost of body condition and increased periparturient disorders in dairy herds. J. Vet. Sci. 7, 161-166. [ Links ]
Lin, C., McAllister, A., Ng-Kwai-Hang, K., Hayes, J., Batra, T., Lee, A., Roy, G., Vesely, J., Wauthy, J. & Winter, K., 1987. Association of milk protein types with growth and reproductive performance of dairy heifers. J. Dairy Sci. 70, 29-39. [ Links ]
Loftus, R., Ertugrul, O., Harba, A., El-Barody, M., MacHugh, D., Park, S. & Bradley, D., 1999. A microsatellite survey of cattle from a centre of origin: the Near East. Mol. Ecol. 8, 2015-2022. [ Links ]
Massella, E., Piva, S., Giacometti, F., Liuzzo, G., Zambrini, A.V. & Serraino, A., 2017. Evaluation of bovine beta casein polymorphism in two dairy farms located in northern Italy. Ital. J. Food Saf. 6, 131-133 [ Links ]
Matejicek, A., Matejickova, J., Nemcova, E., Jandurova, O., Stipkova, M., Bouska, J. & Frelich, J., 2007. Joint effects of CSN3 and LGB genotypes and their relation to breeding values of milk production parameters in Czech Fleckvieh. Czech J. Anim. Sci. 52, 83-87. [ Links ]
Mehta, J.L. & Li, D., 2002. Identification, regulation and function of a novel lectin-like oxidized low-density lipoprotein receptor. J. Am. Coll. Cardiol. 39, 1429-1435. [ Links ]
Meikle, A., Kulcsar, M., Chilliard, Y., Febel, H., Delavaud, C., Cavestany, D. & Chilibroste, P., 2004. Effects of parity and body condition at parturition on endocrine and reproductive parameters of the cow. Reproduction 127, 727-737. [ Links ]
Menezes, M.P.C., Martinez, A.M., Ribeiro, M.N., Pimenta, Filho, E.C. & Bermejo, J.V.D., 2006. Genetic characterization of Brazilian native breeds of goats using 27 markers microsatellites. R. Bras. Zootec. 35, 1336-1341. [ Links ]
Miglior, F., Muir, B.L. & Van Doormal, B.J., 2005. Selection indices in Holstein cattle of various countries. J. Dairy Sci. 88, 1255-1263. [ Links ]
Miluchova, M., Trakovicka, A. & Gabor, M., 2009. Analysis of polymorphism of beta casein of Slovak Pinzgau cattle by PCR-RFLP for allels A1 and A2. Scientific Papers Animal Science and Biotechnologies (SPASB) 42, 288-292. [ Links ]
Mitra, A., Schlee, P., Krause, I., Blusch, J., Werner, T., Balakrishnan, C. & Pirchner, F., 1998a. Kappa-casein polymorphisms in Indian dairy cattle and buffalo: A new genetic variant in buffalo 1. Anim. Biotechnol. 9, 81-87. [ Links ]
Mitra, A. & Yadav, B.R., 1998b. Alpha-lactalbumin polymorphism in three breeds of Indian Zebu cattle. J. Anim. Breed. Genet. 115, 403-405. [ Links ]
Nei, M. & Roychoudhury, A., 1974. Sampling variances of heterozygosity and genetic distance. Genetics 76, 379-390. [ Links ]
Nilforooshan, M. & Edriss, M., 2004. Effect of age at first calving on some productive and longevity traits in Iranian Holsteins of the Isfahan Province. J. Dairy Sci. 87, 2130-2135. [ Links ]
NRC, 2001. Nutrient Requirements of Dairy Cattle. Washington, DC. Natl. Acad. Press, USA. [ Links ]
Oikonomou, G., Angelopoulou, K., Arsenos, G., Zygoyiannis, D. & Banos, G., 2009. The effects of polymorphisms in the DGAT1, leptin and growth hormone receptor gene loci on body energy, blood metabolic and reproductive traits of Holstein cows. Anim. Genet. 40, 10-17. [ Links ]
Olsen, H.G., Nilsen, H., Hayes, B., Berg, P.R., Svendsen, M., Lien, S. & Meuwissen, T., 2007. Genetic support for a quantitative trait nucleotide in the ABCG2 gene affecting milk composition of dairy cattle. BMC Genet. 8, 32-40. [ Links ]
Penagaricano, F. & Khatib, H., 2012. Association of milk protein genes with fertilization rate and early embryonic development in Holstein dairy cattle. J. Dairy Res. 79, 47-52. [ Links ]
Pryce, J., Royal, M., Garnsworthy P. & Mao, I.L., 2004. Fertility in the high-producing dairy cow. Livest. Prod. Sci. 86, 125-135. [ Links ]
Ray, D., Halbach, T. & Armstrong, D., 1992. Season and lactation number effects on milk production and reproduction of dairy cattle in Arizona! J. Dairy Sci. 75, 2976-2983. [ Links ]
Royal, M., Pryce, J., Woolliams, J. & Flint, A., 2002. The genetic relationship between commencement of luteal activity and calving interval, body condition score, production, and linear type traits in Holstein-Friesian dairy cattle. J. Dairy Sci. 85, 3071-3080. [ Links ]
Ruottinen, O., Ikonen, T. & Ojala, M., 2004. Associations between milk protein genotypes and fertility traits in Finnish Ayrshire heifers and first lactation cows. Livest. Prod. Sci. 85, 27-34. [ Links ]
Rychtarova, J., Sztankoova, Z., Kyselova, J., Zink, V., Stipkova, M., Vacek, M. & Stolc, L., 2014. Effect of DGAT1, BTN1A1, OLR1, and STAT1 genes on milk production and reproduction traits in the Czech Fleckvieh breed. Czech J. Anim. Sci. 59, 45-53. [ Links ]
Schennink, A., Bovenhuis, H., Leon-Kloosterziel, K.M., Van Arendonk, J.A. & Visker, M.H., 2009. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Anim. Genet. 40, 909-916. [ Links ]
Sewalem, A., Miglior, F., Kistemaker, G., Sullivan, P. & Van Doormaal, B., 2008. Relationship between reproduction traits and functional longevity in Canadian dairy cattle. J. Dairy Sci. 91, 1660-1668. [ Links ]
Silva, H., Wilcox, C., Thatcher, W., Becker, R. & Morse, D., 1992. Factors affecting days open, gestation length, and calving interval in Florida dairy cattle. J. Dairy Sci. 75, 288-293. [ Links ]
Strzalkowska, N., Krzyzewski, J., Zwierzchowski, L. & Ryniewicz, Z., 2002. Effects of kappa-casein and beta-lactoglobulin loci polymorphism, cows' age, stage of lactation and somatic cell count on daily milk yield composition in Polish Black-and-White cattle. Anim. Sci. Pap. Rep. 20, 21-35. [ Links ]
Tekerli, M. & Kocak, S., 2009. Relationships between production and fertility traits in first lactation and life time performances of Holstein cows under subtropical condition. Arch. Tierz. 52, 364-370. [ Links ]
Thaller, G., Kühn, C., Winter, A., Ewald, G., Bellmann, O., Wegner, J., Zühlke, H. & Fries, R., 2003. DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Anim. Genet. 34, 354-357. [ Links ]
Tozer, P. & Heinrichs, A., 2001. What affects the costs of raising replacement dairy heifers: A multiple-component analysis. J. Dairy Sci. 84, 1836-1844. [ Links ]
Trakovicka, A., Moravcikova, N. & Kasarda, R., 2013. Genetic polymorphisms of leptin and leptin receptor genes in relation with production and reproduction traits in cattle. Acta Biochim. Pol. 60, 783-787. [ Links ]
Tsiaras, A., Bargouli, G., Banos, G. & Boscos, C., 2005. Effect of kappa-casein and beta-lactoglobulin loci on milk production traits and reproductive performance of Holstein cows. J. Dairy Sci. 88, 327-334. [ Links ]
Yeh, F.C., Yang, R.C., Boyle, T.B., Ye, Z. & Mao, J.X., 2000. POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada. [ Links ]
Received 27 March 2018
Accepted 13 March 2019
First published online 8 May 2019
# Corresponding author: fbalci@uludag.edu.tr














