Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.47 n.4 Pretoria 2017
http://dx.doi.org/10.4314/sajas.v47i4.7
ARTICLES
Evaluation of production and reproduction of three South African Angora goat CYP17 genotypes
M.A. SnymanI, #; K-H. StorbeckII; P. SwartII
IGrootfontein Agricultural Development Institute, Private Bag X529, Middelburg (EC), 5900
IIDepartment of Biochemistry, University of Stellenbosch, Private Bag X1, Matieland, 7602
ABSTRACT
Two CYP17 genes, located on different loci and expressing enzymes with significantly different activities, have been identified in the South African Angora goat population. Three unique genotypes (named He, Hu, and Ho), which differed not only in the genes encoding CYP17, but also in copy number were subsequently identified in the Angora goat. The aim of this study was to evaluate the production and reproduction performance of these three genotypes. Bodyweight, fleece and reproduction data, and blood samples from 466 Angora ewes from three flocks were obtained from the GADI-Biobank. Data had been collected on Flock 1 from 2000 to 2015, Flock 2 from 2000 to 2014 and Flock 3 from 2000 to 2010. Bodyweight data included birth weight, weaning weight, 8-, 12-, and 16-month bodyweight, as well as bodyweight recorded annually for the ewe flock before mating. Fleece data included fleece weight and fibre diameter recorded at the second and third shearings at 12 and 18 months old, respectively. Fleece weight, fibre diameter, style and character were also recorded annually for the ewe flocks during the winter shearing. Individual reproduction records included information on whether the ewe had kidded, whether the ewe had aborted, number of kids born, stillborn kids, kids that died soon after birth, kids reared by a foster mother, kids reared as orphans, number of kids weaned and total weight of kids weaned. Total lifetime reproductive performance of genotyped ewes was calculated for number of kids born, number of kids weaned, and total weight of kids weaned. Blood samples were also collected from 100 sexually active Angora rams from four sources. CYP17 genotyping was carried out using an ARMS-qPCR (amplification refractory mutation system qPCR) assay. Serum testosterone was quantified using high performance liquid chromatography mass spectrometry. The distribution of the ewes across the three CYP17 genotypes was 36.7% He, 51.5% Hu, and 11.8% Ho, and was in accordance with the distribution of the Angora veld rams (38.0% He, 46.4% Hu, and 15.6% Ho). In this study, animals of the Hu genotype were heavier from weaning age onwards, although this difference in bodyweight was significant only at 8 months old and in the adult ewes. No differences were observed between the He and Ho animals. Adult ewes of the He genotype (1.35 kg) produced heavier (P <0.05) fleeces than the Hu (1.27 kg) and Ho (1.24 kg) genotypes. Fibre diameter of the fleeces of the Ho ewes (33.0 µm) was higher than that of the fleeces of the He ewes (32.2 µm; P <0.05) and the Hu ewes (32.7 µm; P >0.05). No significant differences were recorded in reproductive performance among the genotypes, although the Ho ewes had the lowest (1.03 and 0.89) and the He ewes the highest (1.07 and 0.93) number of kids born and weaned per year respectively. Results on the rams indicated that the CYP17 genotype had no effect on testosterone production by Angora rams. From the results of this study no evidence could be found that selection for any of the three genotypes would adversely affect any growth, mohair production or reproduction function of Angora ewes. A breeding strategy incorporating selection for productive traits and the CYP17 genotype, aimed at increasing the frequency of the ACS+ gene and thus the He genotype in the population, could be followed without having a negative effect on the genetic progress of productive traits.
Keywords: Bodyweight, fleece weight, number of kids weaned, testosterone, weight of kids weaned
Introduction
One of the continual problems that face the South African mohair industry is the severe loss of Angora goats during cold spells, owing to the inability of the goats to cope with severe stress (Wentzel et al., 1979; Fourie, 1984; Storbeck et al., 2007; Snyman & Van Heerden, 2011; Snyman et al., 2017). Previous research has shown that the Angora goat is unable to produce sufficient cortisol (the adrenal steroid hormone released in response to physiological stress) in reaction to cold stress (Van Rensburg, 1971; Herselman & Pieterse, 1992; Herselman & Van Loggerenberg, 1995; Engelbrecht et al., 2000; Engelbrecht & Swart, 2000; Storbeck et al., 2008; Storbeck et al., 2009). This deficiency in cortisol production could be attributed to anomalies in the adrenal steroidogenesis of the Angora goat. A single enzyme, namely cytochrome P450 17α-hydroxylase/17,20 lyase (CYP17), was identified as the probable cause of the problem (Storbeck et al., 2008). Depending on CYP17 activity, the adrenal steroid hormone biosynthesis pathway is either directed to the formation of mineralocorticoids and glucocorticoids or to precursors of sex hormones (Gilep et al., 2011).
Two CYP17 genes have been identified in the South African Angora goat population, namely ACS-(GenBank accession no. EF524063) and ACS+ (GenBank accession no. EF524064) (Storbeck et al., 2008). The sequences of these two CYP17 genes differ by four single nucleotide polymorphisms and express enzymes with significantly different activities (Slabbert, 2003; Storbeck et al., 2007; Storbeck et al., 2008; Storbeck et al., 2009). Research involving copy numbers of these genes indicated that in both the South African Angora goat and the Boer goat, CYP17 ACS- and ACS+ are not two alleles of a single CYP17 gene, as initially indicated by Slabbert (2003) and Storbeck et al. (2007), but are separate genes that occur at different loci (Storbeck et al., 2008). To date, the Angora and Boer goats are the only mammals to be reported to possess two CYP17 genes encoding two CYP17 isoforms (Nakajin et al., 1981; Chung et al., 1987; Givens et al., 1994). The product of the CYP17 ACS- was shown to exhibit significantly enhanced 17,20-lyase activity which prevented the accumulation and availability of cortisol precursors during adrenal steroidogenesis (Storbeck et al., 2008). This 'abnormal' enzyme activity is believed to be the leading cause of the observed hypocortisolism in these animals (Storbeck et al., 2009). Interestingly, CYP17 alleles of the same gene have been associated with different physiological cortisol responses in Merino sheep (Hough et al., 2010; Hough, 2012; Qui, 2015).
Three unique genotypes, which differed not only in genes encoding CYP17, but also in copy number, have subsequently been identified in the Angora goat (Storbeck et al., 2008). For the purposes of this study these genotypes were denoted He, Huand Ho. The Hogenotype has only one CYP17 gene, namely ACS-. The He genotype has both CYP17 genes (ACS+ and ACS-) at two loci and therefore twice the copy number of Ho with a ACS- : ACS+ ratio of 2 : 2 (Storbeck et al., 2008). Crossing Ho and He goats has been shown to yield an intermediate genotype, Hu, which receives both the ACS- and ACS+ genes from the Heparent, but only the ACS- gene from the Ho parent. Therefore, in this genotype, the ACS- : ACS+ ratio would be 2 : 1, which is supported by the copy number, where the Hugenotype yielded a 1.4-fold greater copy number than the Hogroup (Storbeck et al., 2008). Although the location of the ACS- and ACS+ genes has not yet been mapped in the Angora goat, data obtained from preliminary breeding studies have confirmed the existence of these three genotypes (Swart, 2011).
An insulin-induced stress experiment confirmed that there was a significant difference in the ability of the three genotypes to produce cortisol in response to physiological stress (Storbeck et al., 2008; Hough et al., 2010). The He genotype was the best cortisol producer during the insulin-induced stress test, and the Ho genotype produced the least cortisol. Subsequent to the insulin-induced stress test, a simulated cold stress test was performed at Grootfontein Agricultural Development Institute (GADI) (Snyman et al., 2017). While the Hogenotype again produced the least cortisol during the cold stress test, the rectal temperature of the Hugenotype dropped significantly more than that of the other two genotypes. The He genotype was the best performer in both the insulin-induced and cold stress tests, and therefore represents the hardiest genotype.
In addition to being important in cortisol production, the CYP17 enzyme is vital for the production of oestrogens (female sex hormones) and androgens (male sex hormones) (Payne & Hales, 2004). The effect of the CYP17 genotypes on the reproductive fitness of Angora ewes was thus investigated (Swart, 2012; Swart, 2013). The results indicated that the Ho genotype is a poor reproducer; 62.5% of the Ho group were poor reproducers, while the Hu and He groups contained 35% and 42% poor reproducers, respectively. Poor reproducers were defined as ewes that had weaned fewer kids than they had had kidding opportunities in the flock. However, a larger sample group needs to be investigated before final recommendations can be made to the industry.
This study formed part of a bigger project, which investigated the possibility of breeding more hardy Angora goats without negatively affecting the reproductive fitness of the goats or decreasing the quality of mohair, for which South Africa is known. Data gathered to date suggest that the Hegenotype is the hardiest and should be selected for. However, there is a dearth of information of the relative production and reproduction performance of the three genotypes. The aim of this study was therefore to evaluate the production and reproduction performance of the three Angora goat CYP17 genotypes to determine whether selection for the Hegenotype would have a detrimental effect on productive performance and ultimately to be able to make recommendations to the industry about the way forward regarding implementation of selection practices incorporating the CYP17 genotype.
Material and methods
Resources from the GADI-Biobank, including blood samples and phenotypic data, were used for the first part of this study. Reproduction and production data of the Angora ewes from the three flocks in the GADI-Biobank that had the most complete datasets were used. Lifetime reproductive performance of the ewes in each of these flocks was calculated. Ewes with incomplete records for reproduction, bodyweight and hair production and those without available blood samples were omitted. Subsequently, for each flock, ewes from among the good, average and poor reproductive performers were randomly selected to genotype the CYP17 locus. The number of ewes being mated annually in these flocks was approximately 600, 100, and 300. Therefore, the same ratio, as far as possible, was applied to selecting samples for genotyping from the three flocks. Samples from 480 ewes were ultimately selected for genotyping. Blood samples from the ewes were obtained from the GADI-Biobank.
The three flocks were representative of animals across the Angora goat farming spectrum, and represented management systems from very extensive to fairly intensive. For details of management systems followed in the studs, see Snyman (2007; 2010a; 2010b), in which the current Flocks 1, 2, and 3 correspond to Studs 5, 7 and 12 in Snyman (2007; 2010a; 2010b). The average number of kids born per ewe mated in the three flocks was 99%, 72%, and 122% for Flocks 1, 2 and 3, respectively (Snyman, 2010a). The corresponding number of kids weaned per ewe mated was 78%, 58%, and 105%. Adult bodyweights of the ewes in the flocks were 37.0 kg, 33.4 kg, and 44.5 kg. Flock 2 was the only flock in which purposeful selection was carried out for decreased fibre diameter, while maintaining bodyweight and fleece weight, based on performance data (Snyman, 2002). Selection in the other two flocks was mostly done subjectively and was not based on performance data.
In addition to evaluating the reproductive performance of the ewes, testosterone production of Angora rams from the three genotypes was compared. For this part of the study, blood samples were collected from 100 sexually active Angora rams from the farms of three producers (Flock 1, Flock 4, and Flock 5). The rams had all been with the ewes until the week in which the blood samples were collected. Blood samples were also collected from 192 two-tooth Angora rams, which were to be sold at the annual Angora veld ram auctions in January 2013. These rams had been run under the same conditions on the veld since July 2012 and had not been exposed to ewes.
Genomic DNA was isolated from 200 µl blood from all the blood samples using the Quick-DNA™ Universal Kit (ZYMO Research). The extracted DNA samples were used to genotype the CYP17 locus. Genotyping was carried out using an ARMS-qPCR assay. Primer and probe sequences were as follows:
ACS LP, 5'-ACAATGTGGGTGCTCTTG G-3'
ACS- RP, 5'-CACGTCTGGGGAGGAACG-3'
ACS+ RP, 5'-CACGTCTGGGGAGGAACA-3'
ACS TaqMan Probe, FAM-AAGACCAAGCACTCTGCTGCCAAGTA-BHQ
qPCR reactions (20 µl) contained 500 nM of each primer (ACS LP and ACS- RP or ACS LP and ACS+ RP), 500 nM of the TaqMan probe, 1X KAPA PROBE FAST Universal qPCR Master Mix (KAPA biotechnologies) and 5 µl of purified genomic DNA. Amplifications were performed using the LightCycler 96 instrument (Roche Applied Science) with the following cycling conditions: initial denaturation at 95 °C for 3 min, followed by 1 cycle each of 95 °C for 2 sec, 65 °C for 3 sec with 1 °C decrease per cycle for 10 cycles and extension at 72 °C for 2 sec. The reaction was subsequently completed with 35 additional cycles with an annealing temperature of 55 °C. Fluorescence was measured at the end of each extension step (72 °C). A heterozygous (He) calibrator was included in each run. Data were analysed with LightCycler 96 software package (Roche Applied Science). Genotypes of 466 ewes were available for inclusion in the study.
Steroids were extracted from whole blood and quantified by ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS), as described by Schloms et al. (2014).
The following production and reproduction data for the ewes were obtained from the Angora Biobank database of the GADI-Biobank:
- Reproductive performance of ewes
- Bodyweight of ewes before mating
- Fleece weight, fibre diameter, style and character of ewes recorded during the winter shearing
- Bodyweight and fleece data recorded at younger ages
Bodyweight, fleece data and reproduction data had been recorded for Flock 1 from 2000 to 2015, Flock 2 from 2000 to 2014, and Flock 3 from 2000 to 2010. Bodyweight data included birth weight, weaning weight, 8-, 12-, and 16-month bodyweight, as well as bodyweight recorded annually on the ewe flock before mating. Fleece data included fleece weight and fibre diameter recorded at the second and third shearings at 12 and 18 months old, respectively. Fleece weight, fibre diameter, style and character were also recorded annually for the ewe flocks during the winter shearing.
The PROC GLM procedure of SAS (2009) was used to analyse bodyweight and fleece data. The following model was used to analyse the effect of genotype on bodyweight from birth until 16 months old, as well as second and third shearing fleece data:
Yijklmn= µ + hi+ gj+ bk+ rl + am+ b1AGE+ eijklmn
Where: Yijklmn = trait of the nth animal of the mth age of dam of the lth rearing group of the kth birth/rearing status of the jth genotype of the ith flock-year of birth µ = overall mean
hi= fixed effect of the ith flock-year of birth
gj= fixed effect of the jth genotype (He, Ho, Hu)
bk= fixed effect of the kth birth/rearing status (only bodyweights)
rl = fixed effect of the lth rearing group / shearing group
am= fixed effect of the mth age of dam
b1= linear regression coefficient of the age of the animal at recording of the specific bodyweight (only bodyweight)
eijklmn = random error with zero mean and variance Iσ2e
The following model, employing the PROC GLM procedure of SAS (2009), was used to analyse the effect of genotype on adult bodyweight and fleece data:
Yijklm= µ + hi+ gj+ yk+ al+ (hg)ij + (ga)jl+ eijklm
Where: Yijklm= trait of the mth animal of the lth age of the kth year of the jth genotype of the ith flock, µ = overall mean
hi= fixed effect of the ith flock
gj= fixed effect of the jth genotype (He, Ho, Hu)
yk= fixed effect of the kth year of recording
al= fixed effect of the lth age at recording
(hg)ij= effect of the interaction between the ith flock and the jth genotype
(ga)jl= effect of the interaction between the jth genotype and the lth age at recording
eijklm = random error with zero mean and variance Iσ2e
Individual reproduction records included information about whether the ewe had kidded, whether she had aborted, number of kids born, stillborn kids, kids that died soon after birth, kids reared by a foster mother, kids reared as orphans, number of kids weaned and total weight of kids weaned. Total lifetime reproductive performance in the flock for the ewes genotyped was calculated for number of kids born, number of kids weaned and total weight of kids weaned. The same model that had been used for adult bodyweight and fleece data was used to analyse the effect of genotype on individual reproductive performance.
The following model, employing the PROC GLM procedure of SAS (2009), was used to analyse the effect of genotype on flock lifetime reproductive performance:
Yijklm= µ + hi+ gj+ yk+ kl+ eijklm
Where: Yijklm= trait of the mth animal of the lth number of kidding opportunities of the kth year of birth of the jth genotype of the ith flock
µ = overall mean
hi= fixed effect of the itth flock
gj= fixed effect of the jth genotype (He, Ho, Hu)
yk= fixed effect of the kth year of birth of ewe
kl= fixed effect of the lth number of kidding opportunities
eijklm = random error with zero mean and variance Iσ2e
The CHISQ procedure of SAS (2009) was used to determine whether there were differences among the three genotypes in whether the ewe had kidded, whether the ewe had aborted, number of kids born, stillborn kids, kids that died soon after birth, kids reared by a foster mother, kids reared as orphans and number of kids weaned.
The following model, employing the PROC GLM procedure of SAS (2009), was used to analyse the effect of genotype on testosterone level in the Angora rams:
Yijkl= µ + oi+ aj+ gk+ (og)ik+ eijkl
Where: Yijkl = trait of the lth animal of the kth genotype of the jth age of the ith origin
µ = overall mean
oi= fixed effect of the ith origin (Flock 1, 4, 5)
aj= fixed effect of the jth age (4-, 6-, 8-tooth)
gk= fixed effect of the kth genotype (He, Ho, Hu)
(og)ik= effect of the interaction between the ith origin and the kth genotype eijkl= random error with zero mean and variance Iσ2e
For the veld rams, only genotype was included in the model, as all rams were the same age and had been treated as one contemporary group for a seven-month period preceding blood collection.
Results
The distribution of the ewes across the CYP17 genotypes for the pooled data of the ewes and in the three flocks is summarised in Table 1. The highest percentage animals had the Hu genotype in all flocks, as well as when the flocks were pooled. Ho was the least represented, except in Flock 2, where Ho and He were evenly represented.
The fixed effects included in the analyses of the various traits until 16 months old are summarised in Table 2, while those included for the adult bodyweight and fleece traits are presented in Table 3. Genotype had a significant effect on 8-month bodyweight (Table 2), adult bodyweight, adult fleece weight, and style of the fleece (Table 3).
Fixed effects included in the models fitted for reproduction traits are given in Table 4. Genotype had no overall effect on number of kids born, number of kids weaned or total weight of kids weaned. A significant interaction between genotype and flock was observed for adult bodyweight, fleece weight, fibre diameter and the individual reproductive traits.
The effect of genotype on bodyweight from birth until adult age is presented in Table 5. Animals of the Hugenotype were heavier from weaning age onwards, although this difference in bodyweight was only significant at 8 months old and in the adult ewes. No differences were observed between the Heand Hoanimals.
The effect of genotype on the fleece traits from second shearing until adult age is presented in Table 6. No differences were evident at the second or third shearings. Adult ewes of the He genotype (1.35 kg) produced heavier (P <0.05) fleeces than both the Hu (1.27 kg) and Ho (1.24 kg) genotypes. Fibre diameter of the fleeces of the Ho ewes (33.0 µm) was higher (P <0.05) than that of the fleeces of the He ewes (32.2 µm). The fleeces of the Hoewes had the best style.
As already stated, there was a significant interaction between flock and genotype in adult bodyweight (P = 0.009), fleece weight (P = 0.0010) and fibre diameter (P = 0.0336). These interactions are illustrated in Figures 1 to 3, respectively. There was no significant difference in bodyweight among the genotypes in Flock 1, while the Huewes in Flock 2 were heavier than the Heewes (P <0.05) in Flock 2. The Heand Huewes were heavier (P <0.05) than the Ho ewes in Flock 3. He ewes produced the most hair in Flocks 1 and 2 (P <0.05), while He and Hu ewes produced more hair than Ho ewes in Flock 3 (P <0.05) (Figure 2). The biggest differences among genotypes in fibre diameter were recorded in Flock 2
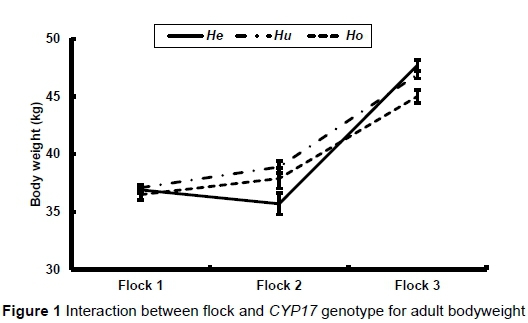
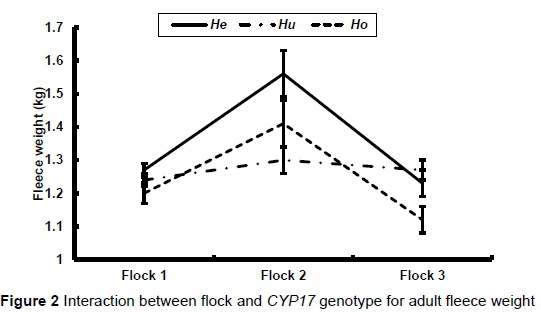
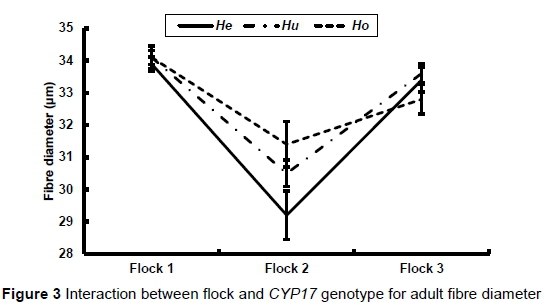
The effect of genotype on the reproductive performance of ewes is presented in Table 7. The Hoand Hegenotypes had the lowest and highest number of kids born and weaned per year, respectively, while Hu ewes had the best lifetime flock reproductive performance. However, none of these recorded differences in reproductive performance among the genotypes were significant.
The effect of genotype on the number of kids born in the various flocks in terms of percentage of ewes of each genotype which gave birth to either 0, 1, 2 or 3 kids is summarised in Table 8. For each flock, within each genotype, the percentage of ewes for all number of kids born categories adds up to 100%.
No significant trend was observed in Flock 1. In Flock 2 approximately 20 to 25% more Ho ewes did not give birth to any kids, compared to the Huand He genotypes, while approximately 20% fewer Hoewes gave birth to 1 kid. In Flock 3 there were no Hoewes that did not produce a kid. Looking at the pooled data, there was no difference among the genotypes in terms of percentage of ewes of each genotype which gave birth to either 0, 1, 2 or 3 kids.
The effect of genotype on the number of kids weaned in the various flocks in terms of percentage of ewes of each genotype which weaned either 0, 1, 2 or 3 kids is summarised in Table 9. There was no significant trend observed in Flock 1, and ewes were spread evenly among genotypes in terms of number of kids weaned category. In Flock 2 more Ho ewes did not wean a kid compared to the He and Hu genotypes. Furthermore, fewer Ho ewes weaned 1 kid and none weaned 2 kids. In Flock 3 fewer Ho ewes weaned 0 kids, while no definite trend was observed among genotype for ewes weaning either 1 or 2 kids. As was the case with number of kids born, again there was no difference in the pooled data among the genotypes in terms of percentage of ewes of each genotype which weaned 0, 1, 2 or 3 kids.
The effect of genotype on the occurrence of ewes that did not kid and ewes that aborted in the various flocks is summarised in Table 10. For these calculations, ewes that did not kid included ewes that aborted. No definite trend could be observed across the flocks. In Flock 1, no differences were recorded among genotypes, while in Flock 2, significantly more Ho ewes did not kid than ewes of the other genotypes. In Flock 3, there were significantly more He ewes that did not kid, while all the Hoewes kidded. In the pooled data, no significant differences among genotypes were recorded. As far as abortions are concerned, no significant differences were observed among the genotypes, although the Huewes had the most abortions in Flocks 2 and 3 and in the pooled data.
The effect of genotype on the occurrence of stillborn kids and kids that died soon after birth is presented in Table 11. No major differences were recorded among genotypes, except for Flock 2 where the only recorded stillborn kids were from ewes of the Hugenotype.
The effect of genotype on the occurrence of kids being fostered or reared as orphans in the various flocks is summarised in Table 12. The highest percentage of kids that had to be fostered was born to Hoewes in Flock 1. Most kids that had to be reared as orphans were also born to Hoewes in Flock 1. All orphans in Flock 3 were born to Heewes.
There was a significant interaction between flock and genotype in the reproductive traits. This interaction for total weight of kids weaned (P <0.0371) is illustrated in Figure 4. There was no significant difference in weight of kids weaned among the genotypes in Flock 1, while Hoewes weaned the lowest weight of kids in Flock 2 (P >0.05) and the Hu ewes weaned the lowest weight of kids (P <0.05) in Flock 3 (Figure 4). The same trends were observed for number of kids born and number of kids weaned.
The distribution of the veld rams across the genotypes is summarised in Table 13. As was the case with the ewes, the highest percentage of the veld rams had the Hu genotype, while the Ho genotype was the least represented. The Hu genotype was also the most abundant among the Flock 5 sires, while the He genotype was the most abundant among the Flock 4 sires. All genotypes were equally represented among the Flock 1 sires.
Testosterone levels of Angora rams from Flocks 1, 4 and 5 and the Angora veld rams are summarised in Table 14 for the three genotypes. No significant differences were observed in testosterone levels between rams of the three genotypes in any of the flocks. There was a significant difference when comparing the testosterone levels of rams from Flock 4 (P <0.01) to those from Flock 1 and Flock 5 (Figure 5).
Testosterone levels of the rams from Flock 4 were approximately 4 times higher than those from Flocks 1 and 5.
Discussion
The distribution of the ewes across the three CYP17 genotypes of 36.7% for He, 51.5% for Hu, and 11.8% for Ho is in accordance with that of the Angora veld rams in which 38.0% rams had a He, 46.4% a Hu, and 15.6% a Ho genotype. The sample size from the other localities was much smaller and could thus not be regarded as representative of the population. However, the high proportion Hosires used in Flock 1 and Flock 4 indicates that a higher proportion of Hosires were selected, as the Hogenotype only comprised 10% of the ewes in Flock 1. The current distribution differs somewhat from earlier genotyping using a different genotyping method (HybProbe method), where 42.9% He, 40.6% Hu, and 16.5% Ho animals were observed (Storbeck et al., 2011). However, this previous method could not always accurately distinguish between He and Hugenotypes. An 80% correlation between the two genotyping methods was observed (Swart, 2011).
In a random mating population with the same number of He, Huand Ho animals, in which all possible matings among the genotypes have an equal opportunity of taking place, four of each possible six matings would yield Hugenotypes as one of the progeny genotypes. Over the years, this could have contributed to the occurrence that the Hu animals are the most abundant in most of the flocks.
In this study, animals of the Hugenotype were heavier from weaning age onwards, although this difference in bodyweight was significant only at eight months old and in the adult ewes. No differences were observed between the Heand Hoanimals. These results are in accordance with preliminary results found for the fine hair goats kept at the Jansenville Experimental Station that were born between 2000 and 2008, which included 117 He, 119 Hu, and 62 Hoanimals. No significant differences in bodyweight were observed among the genotypes for these ewes genotyped with the earlier method (Storbeck et al., 2011). This preliminary study is the only one that is available in which the production performance of Angora CYP17 genotypes was evaluated. No other literature studies could be found that related CYP17 isoforms to production traits in goats or sheep.
In the current study, adult ewes of the He genotype produced the heaviest fleeces with the lowest fibre diameter. In the earlier study on fine hair ewes, no significant differences in any of the fleece traits were observed among the genotypes (Storbeck et al., 2011).
In the current study, no significant differences were recorded in reproductive performance among the genotypes, although the Ho ewes had the lowest (1.03 and 0.89) and the He ewes the highest (1.07 and 0.93) number of kids born and weaned per year, respectively. In a preliminary study, 98 ewes of Flock 1 were genotyped with the method used in the current study. In this earlier study, 62.5% of the Ho ewes were poor reproducers, while the Huand He groups had 35% and 42% poor producers, respectively. Ewes were classified as poor reproducers when they weaned fewer kids than they had kidding opportunities. However, only eight ewes with Hogenotypes were identified in the sample group and there was no significant difference between the Hoand other genotypes (Swart, 2012). Hough (2012) also reported that reproduction of Merino sheep seemed to be unaffected by the CYP17 genotype.
Results of the rams indicated that the CYP17 genotype had no effect on testosterone production by Angora rams. The observation that the testosterone levels of the rams from Flock 4 were approximately four times higher than those from Flocks 1 and 5 could be because Flock 4 rams were kept under kraal conditions on a high protein and high energy diet, while the rams from Flocks 1 and 5 were kept under veld conditions.
Angora stud breeders use different selection indices, in which emphasis is placed on fibre diameter, fleece weight and bodyweight in varying ratios (Snyman et al., 1996; Snyman & Olivier, 1996). Snyman (2002) concluded that selection for decreased fibre diameter, while maintaining or increasing bodyweight and fleece weight, led to genetic improvement of these traits in South African Angora goats. This has led to a decrease in the fibre diameter of the South African clip. The clip composition changed from 51% strong adult mohair, 17% fine adult mohair, 15% young goat mohair and 17% kid mohair in 2001 (Mohair South Africa, 2005) to 33% strong adult mohair, 26% fine adult mohair, 23% young goat mohair and 19% kid mohair in 2015 (SAMGA, 2016). Recently, many breeders have moved away from putting too much emphasis on fibre diameter, arguing that it has reached sufficiently fine levels, to selecting strategies in which more emphasis is placed on fleece weight.
The aim of the current study was to determine if preference were given to a specific genotype, He for example, would there be a detrimental effect on production or reproduction? The results have indicated that there were no significant effects of genotype on production or reproduction, which is a positive finding in this case. Thus it does not matter which CYP17 genotype Angora producers select for. Production and reproduction would not be adversely affected. Selection for production and reproduction traits could continue independently of the selection for a specific CYP17 genotype if producers want to incorporate selection for hardiness into their breeding programme.
The CYP17 genotype exerts its effect on the stress coping ability of the animal via cortisol production in the adrenal cortex. The HeCYP17 genotype was previously identified as the hardiest genotype pertaining to stress coping ability in the Angora goat. For breeders interested in incorporating the CYP17 genotype into their selection strategy, it is recommended that all Hogenotype rams should be culled as a first step. Preferably only Hesires should be used. Because the Hugenotype is in abundance in most of the flocks, it would not be feasible to cull possible Husires with desirable production characteristics. These sires, however, should be mated only to Hegenotype ewes, as these matings would yield only Heand Hugenotype offspring. Mating of Hu sires to Ho ewes would yield Ho and Hu progeny, while a mating between a Hu sire and Hu ewes would yield Ho, Hu and He progeny. It is not possible to get rid of the ACS- gene, as all the animals possess it. An effort, however, could be made to decrease the number of animals that do not have the ACS+ gene (the Ho genotype), thereby increasing the frequency of the ACS+ gene in the population.
Conclusions
From the results of this study, no evidence could be found that selection for any of the three genotypes would adversely affect any growth, mohair production or reproduction function of Angora ewes. The CYP17 genotype had no observable effect on the reproductive fitness of rams, as measured by testosterone production. A breeding strategy could be followed that incorporated selection for productive traits and the CYP17 genotype, aimed at increasing the frequency of the ACS+ gene and thus the Hegenotype in the population, without negatively impacting on genetic progress in the productive traits.
Acknowledgements
Mohair South Africa is acknowledged for funding the project. Angora breeders who participated in the Angora Biobank project are also acknowledged for their contributions.
Authors' contributions
MAS: drafting of project proposal, identification and selection of samples, data preparation and data analyses, writing of manuscript. K-HS: genotyping of samples, assisting with drafting of project proposal and writing of manuscript, final reviewing of manuscript. PS: assisting with drafting of project proposal, reviewing of manuscript. All authors read and approved the final manuscript.
Conflict of Interest Declaration
None of the authors have any conflicts of interest to declare.
References
Chung, B.C., Picado-Leonard, J., Haniu, M., Bienkowski, M., Hall, P.F., Shively, J.F. & Miller, W.L., 1987. Cytochrome P450 C17 (steroid 17a-hydroxylase /17,20-lyase): cloning of human adrenal and testis cDNAs indicates that the same gene is expressed in both tissues. Proc. Natl. Acad. Sci. USA 84, 407-411. [ Links ]
Engelbrecht, Y., Herselman, T., Louw, A. & Swart, P., 2000. Investigation of the primary cause of hypoadrenocorticism in South African Angora goats (Capra aegagrus): A comparison with Boer goats (Capra hircus) and Merino sheep (Ovis aries). J. Anim. Sci. 78, 371-379. [ Links ]
Engelbrecht, Y. & Swart, P., 2000. Adrenal function in Angora goats: A comparative study of adrenal steroidogenesis in Angora goats, Boer goats, and Merino sheep. J. Anim. Sci. 78, 1036-1046. [ Links ]
Fourie, T.J., 1984. 'n Vergelykende studie van die effek van koue blootstelling op die hitteproduksie van Angora (Capra aegagrus) en Boerbokke (Capra hircus). MSc thesis, Universiteit van Port Elizabeth. [ Links ]
Givens, C.R., Zhang, P., Bair, S.R. & Mellon, S.H., 1994. Transcriptional regulation of rat cytochrome P450c17 expression in mouse Leydig MA-10 and adrenal Y-1 cells: Identification of a single protein that mediates both basal and cAMP-induced activities. DNA Cell. Biol. 13, 1087-1098. [ Links ]
Gilep, A.A., Sushko, T.A. & Usanov, S.A., 2011. At the crossroads of steroid hormone biosynthesis: The role, substrate specificity and evolutionary development of CYP17. Biochim. Biophys. Acta , Proteins Proteomics 1814(1), 200-209. [ Links ]
Herselman, M.J. & Pieterse, D.M.E., 1992. Effek van kortisolbehandeling op Angorabokke. Proc. 31st Congr. S. Afr. Soc. Anim. Prod. Zithabizeni, 13-16 April, South Africa. [ Links ]
Herselman, M.J. & Van Loggerenberg, A.A., 1995. Further evidence of hypo-adrenocorticism in Angora goat kids. Proc. 34th Congr. S. Afr. Soc. Anim. Prod. Bloemfontein, 10-13 April, South Africa. [ Links ]
Hough, D., 2012. Comparison of two CYP17 isoforms: Implications for cortisol production in the South African Merino. PhD thesis. University of Stellenbosch. [ Links ]
Hough, D., Cloete, S.W.P., Swart, P. & Storbeck, K.-H., 2010. Improving stress coping ability: comparison between the CYP17 genotype of Ovis aries and Capra hircus. 9th Wrld. Congr. Gen. Appl. Livest. Prod. 1-6 Aug 2010, Leipzig, Germany. [ Links ]
Mohair South Africa, 2005. Mohair Review, 2005. Mohair South Africa. Port Elizabeth. [ Links ]
Nakajin, S., Shively, J.E., Yuan, P., Hall, P.F., 1981. Microsomal cytochrome P450 from neonatal pig testis: Two enzymatic activities (17α-hydroxylase and C17,20-lyase) associated with one protein. Biochemistry 20, 4037-4042. [ Links ]
Payne, A.H. & Hales, D.B., 2004. Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr.. Rev. 25, 947-970. [ Links ]
Qui, Z., 2015. Gene polymorphisms associated with temperament in Merino sheep. PhD thesis. University of Western Australia. [ Links ]
SAS, 2009. SAS Procedures Guide, Version 9.1.3. Cary, NC, SAS Institute Inc. [ Links ]
Schloms, L., Smith, C., Storbeck, K-H., Marnewick, J.L., Swart, P. & Swart, A.C., 2014. Rooibos influences glucocorticoid levels and steroid ratios in vivo and in vitro: A natural approach in the management of stress and metabolic disorders? Mol. Nutr. Food Res. 58, 537-549. [ Links ]
Slabbert, J.T., 2003. Identification of two CYP17 alleles in the South African Angora goat. MSc thesis. University of Stellenbosch. [ Links ]
SAMGA, 2016. Agenda for 73rd Annual General Congress of the South African Mohair Growers' Association, June 2016, Graaff-Reinet. [ Links ]
Snyman, M.A., 2002. Evaluation of a genetically fine mohair producing herd. Small Rumin. Res. 43, 105-113. [ Links ]
Snyman, M.A., 2007. Body weight and growth rate of South African Angora goat kids under different pre- and post-weaning management systems. S. Afr. J. Anim. Sci. 37 (2), 132-141. [ Links ]
Snyman, M.A., 2010a. Influence of body weight, age and management system on reproduction of South African Angora goat does. S. Afr. J. Anim. Sci. 40(1), 41-53. [ Links ]
Snyman, M.A., 2010b. Factors affecting pre-weaning kid mortality in South African Angora goats. S. Afr. J. Anim. Sci. 40 (1), 54-64. [ Links ]
Snyman, M.A., & Olivier, J.J., 1996. Genetic parameters for body weight, fleece weight and fibre diameter in South African Angora goats. Livest. Prod. Sci. 47, 1-6. [ Links ]
Snyman, M.A. & Van Heerden, M., 2011. Can protective coats alleviate the effect of cold, wet and windy conditions on Angora goats? Grootfontein Agric 11(2), 1-18. [ Links ]
Snyman, M.A., Olivier, J.J., Wentzel, D., 1996. Breeding plans for South African Angora goats. Angora Goat Mohair. 38, 23-31. [ Links ]
Snyman, M.A., Storbeck, K. & Swart, P., 2017. Physiological parameters of three South African Angora goat CYP17 genotypes subjected to cold stress. Grootfontein Agric 17(1), 1-10. [ Links ]
Storbeck, K., Snyman, M.A. & Swart, P., 2011. A review of the role of cytochrome P450 17-hydroxylase/17,20 lyase (CYP17) in hypocortisolism in South African Angora goats. Grootfontein Agric 11(2), 24-37. [ Links ]
Storbeck, K., Swart, A.C. & Swart, P., 2009. CYP17 causes hypocortisolism in the South African Angora goat. Mol. Cell Endocrinol. 300, 121-125. [ Links ]
Storbeck, K., Swart, A.C., Slabbert, J.T. & Swart, P., 2007. The Identification of two CYP17 alleles in the South African Angora Goat. Drug Metab. Rev. 39, 467-480. [ Links ]
Storbeck, K., Swart, A.C., Snyman, M.A. & Swart, P., 2008. Two CYP17 genes in the South African Angora goat (Capra hircus). The identification of three genotypes that differ in copy number and steroidogenic output. FEBS. 275, 3934-3943. [ Links ]
Swart, P., 2011. Developing a genetic marker for the identification of more hardy Angora goats. Progress report: Angora goat research advisory committee. Mohair South Africa. Port Elizabeth. [ Links ]
Swart, P., 2012. Developing a genetic marker for the identification of more hardy Angora goats. Progress report: Angora Goat Research Advisory Committee. Mohair South Africa. Port Elizabeth. [ Links ]
Swart, P., 2013. Developing a genetic marker for the identification of more hardy Angora goats. Final report: Angora Goat Research Advisory Committee. Mohair South Africa. Port Elizabeth. [ Links ]
Van Rensburg, S.J., 1971. Reproductive physiology and endocrinology of normal and aborting Angora goats. Onderstepoort J. Vet. Res. 38, 1. [ Links ]
Wentzel, D., Viljoen, K.S. & Botha, L.J.J., 1979. Physiological and endocrinological reactions to cold stress in the Angora goat. Agroanimalia. 11, 19. [ Links ]
Received 15 November 2016
Accepted 9 May 2017
First published online 5 June 2017
# Corresponding author: GrethaSn@daff.gov.za














