Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.46 n.2 Pretoria 2016
http://dx.doi.org/10.4314/sajas.v46i2.9
Determining the frequencies of B1, B2, B3 and E alleles of the CSN1S1 gene and their effects on milk yield and composition in Saanen goats
D. Turhan Dincel; S. Ardicli; H. Samli; F. Balci#
Uludag University, Faculty of Veterinary Medicine, Department of Genetics, TR-16059 Nilufer, Bursa, Turkey
ABSTRACT
The aim of the study was to determine the frequencies of B1, B2, B3 and E alleles of the CSN1S1 gene and their effects on milk yield and composition in the Saanen goat breed. Milk samples were collected to identify milk composition with Fourier transform infrared (FTIR) spectroscopy. The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was used to characterize the alleles of CSN1S1 gene. The allelic frequencies of the B1, B2, B3 and E alleles were 0.927, 0.073, 0.390, and 0.272, respectively. B1 and B2 alleles did not affect milk yield and composition. B3 allele had significant effects on protein, fat, total solid (TS), solid not fat (SNF), casein and lactose percentages, but not on lactose yield. E allele significantly influenced fat and TS percentages of milk in the Saanen goat breed. The protein (3.58%), fat (3.83%), TS (12.06%), SNF (8.60%) and casein percentages (2.80%) were higher in B3/B3 goats than in B3/NB3 (NB3: non-B3) and NB3/NB3 goats. For E allele, NE/NE (NE: non-E) goats displayed higher fat (4.06%) and TS (12.42%) percentages of milk than their E/E and E/NE counterparts. As a result, the potential for improving milk composition by selecting for B3 and E allele may be significant in Saanen goats.
Keywords: Capra hircus, PCR-RFLP, SNP, milk production
Introduction
In ruminants, 80% of milk proteins consist of four types of casein (αS1, β, αS2 and κ) (Ramunno et al., 2004). Milk caseins are the main nutritional source for newborns in placental mammals (Caravaca et al., 2009). AlphaS1-casein (αS1-CN), beta-casein (β-CN), alphaS2-casein (αS2-CN) and kappa-casein (κ-CN) are expressed in goat milk and encoded by CSN1S1, CSN2, CSN1S2 and CSN3 genes, respectively (Martin et al., 2002). These are organised as a cluster in a 250 kilobase (kb) genomic DNA segment in chromosome 6 in goats (Martin et al., 2002).
The goat CSN1S1 gene has ~16785 base pairs (bp), including 1138 bp of exonic regions and 15647 bp of intronic regions with a total similarity with the corresponding bovine sequence of about 57% (Ramunno et al., 2004). It is known that 18 alleles of CSN1S1 gene have been identified so far, which are associated with different expression levels of αS1-CN in milk (Küpper et al., 2010). These alleles can be grouped in four classes: strong alleles (A, B1, B2, B3, B4, B', C, H, L and M) are associated with 3.5 g/L; intermediate alleles (E and I) are associated with 1.1 g/L; weak alleles (F and G) are associated with 0.45 g/L of αS1-CN content; and null alleles (01, 02 and N) are associated with the absence of αS1-CN in milk (Grosclaude et al., 1997; Ramunno et al., 2000; Ibeagha-Awemu et al., 2008; Mastrangelo et al., 2013). As a result, the characterization of the CSN1S1 gene is important owing to its relationship with cheese production and milk-processing properties (Kumar et al., 2007). While the B1, B2 and B3 alleles originated from single nucleotide substitutions responsible for amino acid replacements, the E allele is characterized by the insertion of a DNA segment between the 124th and the 125th nucleotide of the 19th exon (Jansá-Perez et al., 1994; Bevilacqua et al., 2002). The B1, B2 and B3 alleles of CSN1S1 genes are differentiated from A-type (a, G, H, I, 01, 02) alleles with the G-C transversion at the 22nd nucleotide of exon 10; with the T-C transversion at the eighth nucleotide of exon 4; and with G-A transition occurring at the 14th nucleotide of exon 12, respectively (Acc.No: AJ504710) (Cosenza et al., 2008). The E allele consists of the insertion of long interspersed repeated elements (LINE) of 457-458 bp at the 124th nucleotide of exon 19 (Jansá-Perez et al., 1994).
The polymorphisms at αS1-casein locus affect not only the quantity of casein in goat milk, but also its structural and nutritional characteristics and technological properties (Mastrangelo et al., 2013). Characterization of these animals for CSN1S1 variability is important owing to their relationship with milk-processing properties (Kumar et al., 2007). The milk composition and coagulation properties are fairly associated with the the amounts of αS1 -CN (Clark & Sherbon 2000). Lower percentages of total solid (TS), solid not fat (SNF), fat, protein and casein, and a lower coagulation rate were observed in milks that lacked αS1-CN, compared with milk that contained high amounts of αS1-CN in goats (Clark & Sherbon 2000). Milk from EE goats for the CSN1S1 gene had a significantly higher curdling rate than milk from homozygote B-type (B-type: B1, B2, B3, B4, C, L) individuals in the cheese-making process (Caravaca et al., 2011).
Devold et al. (2011) pointed out that the contents of casein were significantly lower in 'null' milks (1.93%) than in milks of the 'strong' group (2.05%) in Norwegian dairy goats. The results obtained by Caravaca et al. (2008) in a study on Murciano-Granadina goats showed that BB genotyped goat for CNS1S1 gene had a higher level of αS1-CN content (8.50 g/L) of milk than EF (6.9 g/L), BF (6.56 g/L) and EE (6.39 g/L) genotyped goats. Another study by these authors indicated that the CSN1S1 genotype did not affect protein, casein and fat concentrations in Murciano-Granadina goats (Caravaca et al., 2009). For milk yield and composition, an investigation of the Sarda breed showed that CSN1S1 BB goats produced significantly higher protein and casein percentages. In addition, αs1-CN was significantly higher in BB and AB Sarda goats than in AF and BF Sarda goats (Balia, 2013). Dominant effects were found for some genotypes for milk yield (AN and BN), fat yield (AN and BE), protein yield (AN and BE), lactose yield (AN), and TS yield (AN). Unfavourable dominance effects were found for protein contents (AB and AN) and TS contents (AB, AN, and AF) in Alpine, Saanen and Toggenburg goats (Vázquez-Flores et al., 2012). Despite the number of studies that determined the frequency of CSN1S1 alleles, investigations into the effects of the CSN1S1 gene on milk yield and composition are limited. Furthermore, there is no published information about the effects of each B-type allele on milk yield and composition.
The objective of this study was therefore to evaluate the frequencies of B1, B2, B3 and E alleles of CSN1S1 gene and determine their effects on milk yield and composition in Saanen goats.
Materials and Methods
The research was carried out on 123 Saanen goats belonging to two herds located in Bursa, Turkey. These herds were reproductively disconnected, so the goats were not related genetically, and were selected at random within the herds. Goats were reared intensively and milked by machine twice a day. Blood and milk samples were collected from selected animals according to ethical procedures. Ethical approval was received from Uludag University (2014-07/02).
In the study, 5 mL blood was collected from the jugular vein using vacutainer tubes containing EDTA as anticoagulant. These blood samples were stored at +4 °C until they arrived at the genetic laboratory of Faculty of Veterinary Medicine-Uludag University. Genomic DNA was extracted from blood by the phenolchloroform method (Powell & Gannon, 2002). The amount and purity of the DNA samples were measured with a spectrophotometer according to the ratio of absorbance at 260 and 280 nm-260/280 (Nanodorp-2000c,Thermo Scientific Inc.). DNA samples were stored at -80 °C until the PCR-RFLP was performed.
Genotyping at the CSN1S1 locus (B1, B2, B3 and E alleles) was performed by the PCR-RFLP method according to Cosenza et al. (2008) and Torres-Vázquez et al. (2008). For the B1, B2, B3 and E alleles of CSN1S1 genes, 311, 310, 231 and 662 bp PCR products were amplified with the established primer sets, respectively (NCBI Acc.No: AJ504710).
For B1, B2, B3 alleles, PCR reaction was prepared in a final volume of 50 μL containing 100 ng/mL genomic DNA, 10 pmol of each primer (B1 allele; F:5' GAAAAGAGAACATGTACTTTG 3', R:5' CATCTTCCTTTTGAATGTACTT 3', B2 allel; F:5'TTCAAATGGAAAAACATTCTCC 3', R:5' GTCAAATGTATAGGTACAGAT 3', B3 allel; F:5' TTAGTTTCCCATTCTTTACTC 3', R:5' GAAGCTCTAACATGATTTGAT 3'), 1.25 U Taq DNA polymerase, 1X Standard Taq Reaction Buffer (10 mM Tris-HCl, 50 mM KCl, pH:8,3), 1.5 mM MgCb, 0.2 mM dNTP (NEB Biolabs Inc.M03200S, N0447S). PCR amplification conditions consisted of an initial step of 97 °C for 2 min, followed by 30 cycles of 94 °C for 45 sec, 55 °C for 45 sec, 72 °C for 45 sec and a final step of 72 °C for 10 min. Differentiation between alleles B1, B2 and B3 was developed by the PCR-RFLP method, according to Cosenza et al. (2008). An 8 μL of each PCR product was digested with MnlI for B1, B2 alleles and DdeI restriction enzyme for B3 alleles at 37 °C for about 16 hours according to their specific prescription. PCR and enzyme digestion products were evaluated by electrophoresis in 2% agarose gel prepared with 0.5X TBE buffer and ethidium bromide. The bands were visualized with the DNr Minilumi imaging system (Cosenza et al., 2008).
The PCR reaction for E allele was prepared in a final volume of 50 μL, containing 100 ng/mL genomic DNA, 10 pmol of each primer (F:5' ATG GGA TTG AAA ATT CCA TGC 3', R:5' ATA CTA CTG GAA TTT AGG TA 3'), 1.25 U Taq DNA polymerase, 1X Standard Taq Reaction Buffer (10 mM Tris-HCl, 50 mM KCl, pH: 8.3), 1.5 mM MgCl2 0.2 mM dNTP (NEB Biolabs Inc. M03200S, N0447S). PCR amplification conditions included an initial step of 94 °C for 3 min, followed for 35 cycles of 94 °C for 50 sec, 59 °C for 50 sec, 72 °C for 50 sec and a final step of 72 °C for 10 min. Similarly to other alleles, 8 μL of each PCR product was digested with Mspl restriction enzyme at 37 °C for about 16 hours according to the prescription. PCR and enzyme digestion products were evaluated by electrophoresis in 2.5% agarose gel, which was prepared with 0.5X TBE buffer and ethidium bromide. The bands were visualized with the DNr Minilumi imaging system (Torres 2008). The milk data (yields and amounts of milk components) were recorded throughout lactation from 67 Saanen goats in Herd 1. After parturition, milk samples were collected in 100 mL sterile sample containers bimonthly during lactation. Samples were transported at +4 °C to the laboratory within two hours (Balia et al., 2013). Total milk yield per lactation was calculated by the Fleischmann method (Trapez II), which was described in International Committee for Animal Recording (ICAR) Guidelines 2014 (Berger & Thomas, 2005). Collected milk samples were analysed for total protein, fat, TS, SNF, total casein and lactose with Fourier transform infrared (FTIR) spectroscopy (MilkoScan™ FT1, Foss Electric, Hillerød, Denmark) (Berget et al., 2010). As the first step of analyses, genotype frequencies were calculated. Data were processed by Popgene v1.32 software to estimate the genotype frequencies and possible deviation from Hardy-Weinberg equilibrium from Saanen goats (Yeh et al., 2000). Statistical analysis was carried out in Minitab 15 statistical software (Minitab 2000) using the general linear model procedure (GLM). The model employed was:
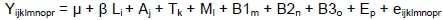
where Yijklmnopris the dependent variable (milk, protein, fat, TS, SNF, casein, lactose yields and rates),
μ is the general mean,
β is the constant for lactation period,
Li is the effect of lactation period (i = 101-288),
Aj is the effect of age (j = 1,2,3,4<),
Tk is the effect of birth type (k = 1,2,3),
Ml is the effect of month of birth (l = February,March),
B1m is the effect of B1 genotype (m = B1,B1/NB1,NB1),
B2n is the effect of B2 genotype (n = B2,B2/NB2,NB2),
B3o is the effect of B3 genotype (o = B3,B3/NB3,NB3),
Ep is the effect of B1 genotype (p = E,E/NE,NE), and
eijklmnopris the random error effect.
For all parameters, model effects were declared significant at P <0.05.
Results and Discussion
The PCR products of B1 allele (311 bp) were digested by restriction enzyme MnlI and showed two fragments for allele B1 at 125 and 186 bp. Allele 'non-B1' (NB1) had a single undigested fragment at 311 bp (Figure 1). B2 homozygote individuals for CSN1S1 gene showed a single undigested fragment of 310 bp, whereas 'non-B2' (NB2) homozygote individuals had two fragments of 77 and 233 bp after the digestion of Mnll restriction enzyme (Figure 2). For B3 allele of CSN1S1 gene single undigested fragment (231 bp) was observed for B3 homozygote individuals, when two fragments of 97 and 134 bp were observed after the digestion of DdeI restriction enzyme for 'non-B3' (NB3) homozygote individuals (Figure 3). A 662 bp fragment was observed in homozygote individuals for E, the 662 and 205 bp fragments were observed in heterozygote individuals, and a 205 bp fragment was observed in homozygote individuals for 'non-E' (NE) alleles (Figure 4).
The genotypic and allelic frequencies of B1, B2, B3 and E alleles at CSN1S1 locus in the Saanen breed are shown at Table 1. The allelic frequencies of the B1, B2, B3 and E alleles were 0.985, 0.030, 0.291 and 0.216 in Herd 1, and 0.857, 0.125, 0.509 and 0.339 in Herd 2, respectively. B1/B1 genotype was the most frequent in Herd 1 and 2 (97.0%, 73.2%). The homozygote 'non-B1 ' genotype was not found in Herd 1, and only one individual was found in Herd 2. For B2 allele, the most frequent genotype was homozygote 'non-B2' with the frequencies of 97.01% and 42% in Herds 1 and 2, respectively. The heterozygote individuals and homozygote individuals for B2 did not exist in Herds 1 and 2. Three genotypes (B3B3, B3/NB3, NB3/NB3) were detected for B3 allele in Saanen goats. Although NB3/NB3 was the most frequent (52.24%) in Herd 1, the most prevalent genotype was determined B3/NB3 (48.21%) in Herd 2. The frequencies of homozygote 'non-E' genotype (62.69%, 44.64%) was determined to be higher than the other genotypes in Herds 1 and 2. In general, B1/B1 (86.18%), NB2/NB2 (86.99%), B3/NB3 (42.88%) and NE/NE genotypes (54.47%) were found to be more frequent in the Saanen breed that was investigated. Although Hardy-Weinberg equilibrium and Χ2-values were not estimated owing to the genotypic frequency for B1 and B2 alleles (n <5 in some groups), significant deviation was not observed for the B3 and E alleles in the breed (P >0.05).
The P-values obtained for each factor of milk yield and composition are given in Table 2. According to the data, unlike the number of kids born (birth type) and month at birth, the protein, fat, total solid, solid not fat, total casein and milk yield were affected by the age and lactation length. Although genotype influenced milk composition, B1 and B2 alleles of the CSN1S1 gene did not affect milk yield and composition (P >0.05). Except for lactose yield, B3 allele has a significant effect on protein, fat, TS, SNF, casein and lactose percentage (P <0.01, P <0.05). The B3 allele tended to be significant on milk yield (P = 0.058). E allele significantly influenced fat and total solid percentages of milk in Saanen goats (P <0.05).
Least square means and pooled standard errors of milk yield and compositions according to the CSN1S1 genotype in the Saanen goat breed are given in Table 3. The protein, fat, TS, SNF and casein percentages were statistical higher in B3/B3 goats than in NB3/NB3 goats. In contrast, the B3/B3 goats had significantly lower lactose percentages than NB3/NB3 goats. Dominance effect was observed for B3 allele for protein, fat, TS, SNF and casein percentages. For fat and TS percentage, NE/NE goats displayed higher fat (4.06%) and TS (12.42%) rates than their E/E counterparts. The fat and total solid percentages were affected by significant dominance effects of the NE allele.
Analysis of the effects of the CSN1S1 gene on goat milk and composition is important to determine the selection scheme. The allelic frequencies of the B1, B2 and B3 alleles were 0.985, 0.030 and 0.291 in Herd 1; 0.857, 0.125 and 0.509 in Herd 2; 0.927, 0.073 and 0.390 in total, respectively. Previous studies showed that the 65 allelic frequencies of B were determined as 0.065 in Girgentana dairy goats (Mastrangelo et al., 2013); 0.05 - 0.230 in Indian goat breeds (Kumar et al., 2007); 0.234 in Bunte Deutsche Edelziege, and 0.103 in Weiße Deutsche Edelziege (Kupper et al., 2010); 0.080 in Hungarian milk goats (Veress et al., 2004); 0.230 in Murciano-Granadina, 0.090 in Malagueña, 0.050 in Alpinee (Jordana et al., 1996); 0.590 in Skopelos (Kalamaki et al., 2014); 0.893 in Czech Fleckvieh (Kučerová et al., 2006); 0.007 in Frisa, 0.008 in Orobica, 0.037 in Verzasca, 0.119 in Camosciata (Caroli et al., 2006); 0.148 in the Maltese goat breed (Chessa et al., 2003). Allelic frequencies of B* were 0.0 (Maga et al., 2014), 0.006 (Grosclaude et al., 1987) and 0.110 (Torres-Vázquez et al., 2008) in Saanen goats, respectively. The allelic frequencies of B1, B2 and B3 alleles (0.927, 0.073, 0.272) in Saanen goats were not similar to data recorded by Cosenza et al. (2008), which was 0.007 for B1, 0.012 for B2 and 0.083 for B3.
The results of the current study showed that the allelic frequencies of E allele were 0.272 in Saanen goats. These results are not consistent with reports in Bunte Deutsche Edelziege (0.171), Weiße Deutsche Edelziege (0.419) (Küpper et al., 2010), Girgentana dairy goats (0.008) (Mastrangelo et al., 2013), Murciano-Granadina (0.590), Malaguena (0.650), Alpinee (0.340) (Jordana et al., 1996), Hungarian milk goats (0.310) (Veress et al., 2004), Maltese (0.057) (Chessa et al., 2003), Skopelos (0.070) (Kalamaki et al., 2014), Camosciata (0.114), Orobica (0.008) (Caroli 2006), White Shorthair (0.054) and Brown Shorthair (0.085) (Sztankóová et al., 2007). Frequencies similar to those of the current study were found only in Frisa (0.200) and Verzasca (0.201) goat breeds by Caroli et al. (2006). In the current study, the frequencies of E were lower than those reported by Soares et al. (2009) (0.350), Grosclaude et al. (1987) (0.410), Torres-Vázquez et al. (2008) (0.420) and Maga et al. (2014) (0.705) in Saanen goats. The diversity of frequencies may be because the goats come from different origins or from herds that were applied different selection parameters.
The present study showed that protein, fat, TS, SNF, casein and milk yield were affected by age and length of lactation. Contrary to results for goats published by Kominakis et al. (2000), lambing month and birth type were not found to be statistically significant for total milk yield. The effects of age on the milk production of goats were similar to data recorded by Mavrogenis et al. (1984). In close agreement with Olechnowicz & Sobek (2008), no significant influence was found for number of kids born on the quantitative parameters of milk yield. The effects of age and lactation length on milk composition should depend on negative correlation with yield and contents of milk.
Balia et al. (2013) pointed out that goats with BB genotypes of the CSN1S1 gene produced higher protein and casein percentages. Unlike the findings of Balia et al. (2013), B1 and B2 alleles of the CSN1S1 gene did not affect milk yield and composition in the present study. On the other hand, significant effects on protein, fat, TS, SNF, casein and lactose percentage were found by B3 allele (P <0.05). So, the positive effects of genotype on milk yield and composition should be derived from B3 alleles compared with B-type. Screening the herds with B3 genotype instead of B-type of CSN1S1 gene may be more felicitous. Altough the protein, fat, TS, SNF, casein percentages were determined to be higher in B3/B3 and B3/NB3 than NB3/NB3 genotypes, lactose percentage was seen to be low in B3/B3 and B3/NB3 genotypes (Table 3). An increase of protein, fat, TS, SNF, casein percentages in B3/B3 and B3/NB3 genotype should occur because of dominant effects of B3 allele. Additionally, B3 allele tended to be significant in the milk yield (P = 0.058). A greater number of animals with copious records should be studied to determine the effect of B3 allele on milk yield. An investigation into Murciano-Granadina goats by Caravaca et al. (2009) indicated that the protein, casein, lactose and SNF percentages were 5.07%, 3.57%, 2.82%, 4.53%, 8.93% in BB genotyped goats, respectively. The results were similar to the authors' data on Saanen goats with B3/B3 genotype shown in Table 3. The fat percentage of milk (5.07%) in Murciano-Granadina (Caravaca et al., 2009) was found to be higher than in the current study of Saanen goats (3.83%). Balia et al. (2013) pointed out that the fat, protein and lactose percentages of milk in Sarda goats with BB genotype was 5.13%, 4.29% and 4.76%, respectively. The fat, protein and lactose percentages of milk in the Saanen goat breed with B3/B3 genotype were found to be lower than specified in Sarda goats by Balia et al. (2013). Similar data were recorded for fat (3.4%) and lactose percentages (4.3%) of milk on Saanen, Alpinee and Toggenburg goats with BB by Flores et al. (2012). Protein (2.9%) and TS percentages (10.7%) were found to be higher than the current data in Saanen with B3/B3.
Caravaca et al. (2009) indicated that fat and SNF percentages were 4.99% and 8.84% in Murciano-Granadina goats with EE genotype. Fat percentage was lower in Saanen goats with EE genotype (as shown in Table 3) than the Murciano-Granadina goat breed (Caravaca et al., 2009). The results of Flores et al. (2012) showed that the fat and total solid percentages of milk in Saanen, Alpine and Toggenburg were 3.18% and 10.79%, respectively. Compared with the study of Flores et al. (2012), fat and TS percentages were higher in Saanen goats with EE and NE/NE genotypes in the present study. NE allele increased the fat and TS percentages of milk statistically in Saanen goats. NE/NE genotype exhibited higher fat and TS percentages than EE and E/NE genotypes. Positive effects on fat and TS percentages of NE should be based on dominant effects.
Conclusion
In conclusion, the effects of B1, B2, B3 alleles of the CSN1S1 gene in Saanen goat breeds were reported for the first time in this publication. According to these results, the allelic frequencies of the B1, B2, B3 and E alleles were 0.927, 0.073, 0.390 and 0.272, respectively. Effects of age and lactation length on protein, fat, TS, SNF, casein and milk yield were found to be significant (P <0.05). However, the environmental factors of month at birth and number of kids born did not affect milk composition. The effects on milk of genotypic factors such as B1 and B2 alleles were not significant in the present study. B3 and E alleles, on the other hand, influenced milk composition considerably (P <0.05). The B3 allele tended to have a significant effect on milk yield (P = 0.058). A greater number of animals with copious records should be studied to clarify the effects of B1, B2, B3 and E alleles on milk yield and composition in Saanen goats. The potential for improving milk composition by selecting for B3 and E allele may be significant. Thus B3 and E alleles could be used as alternative selection criteria for milk composition in Saanen goats. Functional traits such as milk production become important for efficient breeding schemes in dairy goats. In addition, utilizing the genotype information of CSN1S1 gene in selection strategies for milk yield and composition would allow breeders to select production aspects.
Acknowledgements
This study was supported by Uludag University Scientific Research Projects Unit, Project Number OUAP(V)-2011/21.
Authors' contributions
FB coordinated the project design and implementation. DTD and SA was in charge of sample collecting and with HS was responsible for laboratory tests (including DNA isolation and PCR-RFLP procedures). All co-authors participated in results, statistics and interpretation. FB and DTD were in charge of writing the manuscript.
Conflict of interest declaration
We wish to confirm that there are no known conflicts of interest associated with the publication of this manuscript We also confirm that this manuscript has been read and approved by all authors and that the order of authors listed in the manuscript has been approved by all of us.
References
Balia, F., Pazzola, M., Dettori, M.L., Mura, M.C., Luridiana, S., Carcangiu, V., Piras, G. & Vacca, G.M., 2013. Effect of CSN1S1 gene polymorphism and stage of lactation on milk yield and composition of extensively reared goats. J. Dairy Res. 80, 129-137. [ Links ]
Berger, Y.M. & Thomas, D.L., 2005. Milk testing, calculation of milk production and adjustment factors. http://www.ansci.wisc.edu/extensionnew%20copy/sheep/Publications_and_Proceedings/symposium_04. Accessed March 30, 2015. [ Links ]
Berget, I., Martens, H., Kohler, A., Sjurseth, S.K., Afseth, N.K., Narum, B., Ǻdnøy, T. & Lien, S., 2010. Caprine CSN1S1 haplotype effect on gene expression and milk composition measured by Fourier transform infrared spectroscopy. J. Dairy Sci. 93, 4340-4350. [ Links ]
Bevilacqua, C., Ferranti, P., Garro, G., Veltri, C., Lagonigro, R., Leroux, C., Pietrola, E., Addeo, F., Pilla, F., Chianese, L. & Martin, P., 2002. Interallelic recombination is probably responsible for the occurrence of a new as1-casein variant found in the goat species. Eur. J. Biochem. 269, 1293-1303. [ Links ]
Caravaca, F., Amills, M., Jordana, J., Angiolillo, A., Aguera, P., Aranda, C., Mene'Ndez-Buxadera, A., Sànchez, A., Carrizosa, J., Urrutia, B., Sànchez, A. & Serradilla, J.M., 2008. Effect of as1-casein (CSN1S1) genotype on milk CSN1S1 content in malaguenà and Murciano-Granadina goats. J. Dairy Res. 75, 481-484. [ Links ]
Caravaca, F., Carrizosa, J., Urrutia, B., Baena, F., Jordana, J., Amills, M., Badaoui, B., Sanchez, A., Angiolillo, A. & Serradilla, J.M., 2009. Short communication: Effect of αS1-casein (CSN1S1) and K-casein (CSN3) genotypes on milk composition in Murciano-Granadina goats. J. Dairy Sci. 92, 2960-2964. [ Links ]
Caravaca, F., Ares, J.L., Carrizosa, J., Urrutia, B., Baena, F., Jordana, J., Badaoui, B., Sànchez, A., Angiolillo, A., Amills, M. & Serradilla, J.M., 2011. Effects of αs1-casein (CSN1S1) and κ-casein (CSN3) genotypes on milk coagulation properties in Murciano-Granadina goats. J. Dairy Res. 78, 32-37. [ Links ]
Caroli, A., Chiatti, F., Chessa, S., Rignanese, D., Bolla, P. & Pagnacco, G., 2006. Focusing on the goat casein complex. J. Dairy Sci. 89, 3178-3187. [ Links ]
Chessa, S., Ceriotti, G., Dario, C., Erhardt, G. & Caroli, A., 2003. Genetic polymorphisms of αS1-, αS2- and κ-casein in Maltese goat breed. Ital. J. Anim. Sci. 1, 58-60. [ Links ]
Clark, S. & Sherbon, J.W., 2000. Alphas1-casein, milk composition and coagulation properties of goat milk. Small Rumin. Res. 38, 123-134. [ Links ]
Cosenza, G., Pauciullo, A., Gallo, D., Colimoro, L., D'avino, A., Mancusi, A. & Ramunno, L., 2008. Genotyping at the CSN1S1 locus by PCR-RFLP and AS-PCR in a Neapolitan goat population. Small Rumin. Res. 74, 84-90. [ Links ]
Devold, T.G., Nordb, R., Langsrud, T., Svenning, C., Brovold, M.J., S0rensen, E., Christensen, B., Ǻdnøy, T. & Vegarud, G., 2011. Extreme frequencies of the αs1-casein 'null' variant in milk from Norwegian dairy goats-implications for milk composition, micellar size and renneting properties. Dairy Sci. Technol. 91, 39-51. [ Links ]
Grosclaude, F. & Martin, P., 1997. Casein polymorphisms in the goat. Proceedings of an International Seminar: International Dairy Federation milk protein polymorphism seminar II. Bruxelles-Belgium. pp. 241-253. [ Links ]
Grosclaude, F., Mahé, M.F., Brignon, G., Distasio, L. & Jeunet, R., 1987. A Mendelian polymorphism underlying variations of goat as-1-casein. Genet. Sel. Evol.19 (4), 399-412. [ Links ]
Ibeagha-Awemu, E.M., Kgwatalala, P. & Zhao, X., 2008. A critical analysis of production-associated DNA polymorphisms in the genes of cattle, goat, sheep, and pig. Mamm. Genome 19, 591-617. [ Links ]
Jansà-Perez, M.J., Leroux, C., Bonastre, A.S. & Martin, P., 1994. Occurrence of a LINE sequence in the 3' UTR of the goat alpha s1-casein E-encoding allele associated with reduced protein synthesis level. Gene 147, 179-187. [ Links ]
Jordana, J., Amills, M., Diaz, E., Angulo, C., Serradilla, J.M. & Sánchez, A., 1996. Gene frequencies of caprine αs1-casein polymorphism in Spanish goat. breeds. Small Rumin. Res. 20, 215-221. [ Links ]
Kalamaki, M.S., Gelasakis, A. & Arsenos, G., 2014. Genetic polymorphism of the CSN1S1 gene in the Greek-indigenous Skopelos goat. Proceedings of the 4th ISOFAR Scientific Conference: 'Building Organic Bridges', at the Organic World Congress, 13-15 October 2014. pp. 575-577. [ Links ]
Kominakis, A., Rogdakis, E., Vasiloudis, C. & Liaskos, O., 2000. Genetic and environmental sources of variation of milk yield of Skopelos dairy goats. Small Rumin. Res. 36, 1-5. [ Links ]
Kučerová, J., Matějíček, A., Jandurová, O.M., Sørensen, P., Němcová, E., Štípková, M., Kott, T., Bouška, J. & Frelich, J., 2006. Milk protein genes CSN1S1, CSN2, CSN3, LGB and their relation to genetic values of milk production parameters in Czech Fleckvieh. Czech J. Anim. Sci. 51 (6), 241-247. [ Links ]]
Kumar, A., Rout, P.K., Mandal, A. & Roy, R., 2007. Identification of the CSN1S1 allele in Indian goats by the PCR-RFLP method. Animal 1, 1099-1104. [ Links ]
Küpper, J., Chessa, S., Rignanese, D., Caroli, A. & Erhardt, G., 2010. Divergence at the casein haplotypes in dairy and meat goat breeds. J. Dairy Res. 77, 56-62. [ Links ]
Maga, E.A., Daftari, P., Kultz, D. & Penedo, M.C.T., 2009. Prevalence of αs1-casein genotypes in American dairy goats. J. Anim. Sci. 87, 3464-3469. [ Links ]
Martin, P., Szymanowska, M., Zwierzchowski, L. & Leroux, C., 2002. The impact of genetic polymorphisms on the protein composition of ruminant milks. Reprod. Nutr. Dev. 42, 433-459. [ Links ]
Mastrangelo, S., Sardina, M.T., Tolone, M. & Portolano, B., 2013. Genetic polymorphism at the CSN1S1 gene in Girgentana dairy goat breed. Anim. Prod. Sci. 53, 403-406. [ Links ]
Mavrogenis, A.P., Constantinou, A. & Louca, A., 1984. Environmental and genetic causes of variation in production traits of Damascus goats. 2. Goat productivity. Anim. Prod. 38, 99-104. [ Links ]
Olechnowicz, J. & Sobek, Z., 2008. Factors of variation influencing production level, SCC and basic milk composition in dairy goats. J. Anim. Feed Sci. 17, 41-49. [ Links ]
Powell, R. & Gannon, F., 2002. Purification of DNA by Phenol Extraction and Ethanol Precipitation. Oxford University Press 17, 1-2. [ Links ]
Ramunno, L., Cosenza, G., Pappalardo, M., Pastore, N., Gallo, D., Di Gregorio, P. & Masina, P., 2000. Identification of goat CSN1S1F allele by means of PCR-RFLP method. Anim. Genet. 31, 333-346. [ Links ]
Ramunno, L., Cosenza, G., Rando, A., Illario, R., Gallo, D., Berardino, D. D. & Masina, P., 2004. The αs1-casein gene: gene structure and promoter analysis. Gene 334, 105-111. [ Links ]
Soares, M.A.M., Rodrigues, M.T., Mognol, G.P., Ribeiro, L.F.C., Silva, J.L.C. & Brancalhäo, R.M.C., 2009. Polymorphism of alphas1-casein gene in a dairy goat herd in the southeastern region of Brazil. Rev. Bras. Zootecn. 38 (6), 1026-1032. [ Links ]
Sztankóová, Z., Mátlová, V. & Malá, G., 2007. Genetic polymorphism at the CSN1S1 gene in two Czech goat breeds. Czech J. Anim. Sci. 52 (7), 199-202. [ Links ]
Torres-Vázquez, J.A., Vázquez Flores, F., Montaldo, H.H., Ulloa-Arvιzu, R., Posadas, M.V., Vázquez, A.G. & Alonso-Morales, R.A., 2008. Genetic polymorphism of the αs1-casein locus in five populations of goats from Mexico. Electron. J. Biotechnol. 11 (3), 2-11. [ Links ]
Vázquez Flores, F., Montaldo, H.H., Torres-Vázquez, J.A., Alonso-Morales, R.A., Gayosso-Vázquez, A., Valencia-Posadas, M. & Castillo-Juárez, H., 2012. Additive and dominance effects of the αs1-casein locus on milk yield and composition traits in dairy goats. J. Dairy Res. 79, 367-374. [ Links ]
Veress, G., Kusza, S., Bösze, Z., Kukovics, S. & Jávor, A., 2004. Polymorphism of the αs1-casein, κ-casein and β-lactoglobulin genes in the Hungarian Milk Goat. S. Afr. J. Anim. Sci. 34, 20-23. [ Links ]
Yeh, F.C., Yang, R.C., Boyle, T.B.J., Ye, Z.H. & Mao, J.X., 2000. Popgene (v.1.32) the user-friendly shareware for population genetic analysis University of Alberta, Canada: http://www.ualberta.ca/~fyeh/, Accessed January 01, 2015. [ Links ]
Received 20 April 2015
Accepted 3 April 2016
First published online 4 June 2016
# Corresponding author: fbalci@uludag.edu.tr














