Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Animal Science
versión On-line ISSN 2221-4062
versión impresa ISSN 0375-1589
S. Afr. j. anim. sci. vol.44 no.1 Pretoria ene. 2014
SHORT COMMUNICATION
Effect of polymorphism in the LIF gene on reproductive performance of hybrid Polish Large White and Polish Landrace sows
D. Napierala; M. Kawęka; E. Jacyno; B. Matysiak; A. Wierzchowska
Department of Pig Breeding, Animal Feeding and Food, Faculty of Biotechnology and Animal Science, West Pomeranian University of Technology in Szczecin, Poland
ABSTRACT
The aim of the study was to determine the potential relationships between variants of the leukaemia inhibitory factor (LIF) gene and litter size in Landrace x Polish White sows. To identify polymorphisms within the LIF gene, the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method was applied, using specific primers and the DralII enzyme. The researchers identified the presence of two alleles, T and C, with frequencies 0.64 and 0.36, giving three genotypes with frequencies of 0.44, 0.41 and 0.15, respectively, for TT, CT and CC. Analysis of relationships among the various genotypes of the LIF gene and selected reproductive traits showed differences. Sows with genotypes TT and TC in the LIF locus gave birth and raised significantly more live piglets in the first litter compared with sows with the CC genotype.
Keywords: Leukaemia inhibitory factor gene, litter size, pig, reproduction, SNP
One of the greatest challenges faced by pig farming is to find effective tools to improve the reproductive performance of pigs, which is one of the main factors determining the profitability of pig farming (Buske et al., 2006). The reproductive performance of sows is described by many traits, the most significant being litter size, that is, the number of piglets born and raised in the litter, which has a direct impact on the cost of pork production (Spötter & Distl, 2006; Mucha et al., 2013). In addition, one sow should give at least four litters per year to optimize production (Rodriguez-Zas et al., 2006). A consequence of the polygenicity of reproductive traits is that breeding value is often influenced by a large number of genes with various effects on traits important to breeders. Owing to the low level of heritability of reproductive traits (≈0.1), traditional breeding methods based on selection are time consuming and not efficient (Tyra & Różycki, 2004). An alternative to traditional methods of breeding is an analysis using a molecular basis (Short et al., 1997). Spötter et al. (2005) indicated the gene encoding the leukaemia inhibitory factor (LIF) as a potential genetic marker that affects one of the main traits of reproductive performance, litter size. LIF is a cytokine with pleiotropic action that is involved in many physiological processes, including proliferation, differentiation and cell survival. Owing to the key role of LIF in growth and blastocyst implantation in mice (Stewart, 1994), the LIF gene was selected as a potential candidate gene for litter size. In 2006 a study was published that suggested a relation between mutations in the LIF gene and infertility in women (Králíčková et al., 2006). A recent report (Ropka-Molik et al., 2012) on the LIF gene confirms its role in preparing the uterus for the implantation of the embryo in pigs.
The aim of the research was to determine the frequency of particular genotypes of the LIF gene and potential relationships between genotypes of the LIF gene and litter size in hybrid sows.
In this study 493 Polish Large White x Polish Landrace sows, which gave 1760 litters, were used. All animals were bred and raised on a commercial farm (West Pomeranian Region, Poland, EU). Rearing and feeding conditions were identical for all of the animals. All litters were weaned on day 30 of lactation. Sows were kept in a closed house in groups. They were fed a complete feed mixture containing cereals (wheat, barley, triticale, maize, wheat bran) and extracted soybean meal, according to standard in Poland (1993).
During gestation, from days 1 to 90, the sows consumed 2.3 kg of diet/day with a nutritive value of 11.5 MJ ME, 126 g total protein, 5.5 g lysine, 3.4 g methionine with cystine, 7.5 g Ca and 4.8 g P per kg. From 91 to 110 days of gestation they consumed on average 3.0 kg of the diet/day, and during lactation 6.0 kg/day of a diet with a nutritive value of 13.2 MJ ME, 172 g total protein, 9.5 g lysine, 6.8 g methionine with cystine, 8.5 g Ca and 6.5 g P per kg. Water was provided by nipple drinkers. The sows were inseminated with semen obtained from boars from the Pig Improvement Company (PIC).
A blood sample (5 mL) was collected from the jugular veins of sows on a single day, 4 h after feeding (Ethic Committee approval nr 31/2007). The collected material was stored in a vacuum test tube containing K3EDTA (acting as anticoagulant) and transported to the laboratory of the Department of Pig Breeding, Animal Feeding and Food at West Pomeranian University of Technology, Szczecin, Poland. Extraction of genomic DNA was performed with the Master PureTM Genomic DNA Purification Kit of Epicentre Technologies® (Madison, WI, USA). Spectrophotometric analysis showed a mean concentration of DNA within 70 - 90 µg/mL and purity of 90%. In addition, 0.7% agarose gel electrophoresis confirmed the integrity of all DNA samples.
The DNA samples were stored at -20 °C until further analysis. The genotypes of the LIF gene were identified by means of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The primer sequences were 5' ATG TGG ATG TGG CCT ACG G 3' for the forward primers and 5' GGG AAC AAG GTG GTG ATG G 3' for the reverse primers (GenBank: AJ296176.1). Detection of LIF polymorphism was based on the method reported by Spotter et al. (2001). The PCR reactions were performed in a thermocycler (Biometra T Gradient Thermocycler) using thermal profiles adapted to the melting point of each pair of primers and the length of amplicons obtained. The reaction mixture contained 70 ng DNA, 10 pM of each primer, and the samples were amplified using the reaction mixture REDTaq® Ready MixTM (Sigma). Primer sequences were synthesized by the Institute of Biochemistry and Biophysics Polish Academy of Sciences (Warsaw, Poland).
Evaluation of the products obtained after digestion using the DraIII (AdeI) enzyme (MBI Fermentas Burlington, Ontario, Canada) (PCR-RFLP) performed by agarose gel electrophoresis (PRONA) at a concentration matched to the length of the DNA fragments by using the PUC19 DNA/MSPI ladder (MBI Fermentas Burlington, Ontario, Canada), with the addition of ethidium bromide (Table 1). Polymorphism of LIF was detected with a UV transilluminator (Vilber Lourmat).
Data regarding the litter size of sows in subsequent reproductive cycles and weaning-fertile mating intervals were collected, based on breeding records. The characteristics of the genetic structure of the tested sows included allele frequencies and genotypes of LIF, and comparisons of genotype frequencies and genotypes LIF homo- and heterozygous gene LIF from the expected frequencies. Hardy-Weinberg equilibrium in the studied herd was tested by comparing the observed and the expected distributions of genotypes using the chi-squared test (χ2). In order to determine the presence or absence of dependencies between LIF gene variants and the phenotypic values of the analyzed traits, analysis of variance was performed using GLM (generalized linear model) and GLZ multivariate regression methods according to the following model:
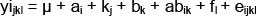
yijkl:phenotypic record of NBA (number of piglets born alive), NSB (number of stillborn piglets), NW (number of piglets weaned)
µ: general mean,
ai: effect of genotype kj: random effect of sire
bk: effect of parity number (j = 1,2,...)
abik: effect of interaction of genotype x parity number
fl: effect of season of birth
eijkl: random error.
The significance of differences between the values of selected characteristics of reproductive performance of sows of different genotypes of the LIF gene was determined using multiple Duncan tests (Duncan, 1955). All analyses were performed using Statistica PL 10 (StatSoft Inc., 2010).
In the analysed herd of sows, two alleles, C and T, of the LIF gene's polymorphic site were identified, and consequently three genotypes, CC, TC and TT.
The study showed (Table 2) a higher presence of allele T (0.64) compared with allele C (0.36), which resulted in a relatively high frequency of the TT genotype (0.44) compared with the second homozygous genotype, CC (0.15), while the heterozygous genotype, CT, had a frequency of 0.41. Additionally, homozygous genotypes were more frequent (0.59) than heterozygous ones (0.41).
The χ2 test showed that there were differences (P <0.05) between the observed and estimated frequencies of LIF genotypes in the sows, and there was a genetic imbalance between the expected and observed number of homo- and heterozygous genotypes of the LIF gene, with the differences being confirmed statistically significant (P <0.05).
These frequencies of alleles and genotypes are consistent with the results reported by Spötter et al. (2005), who conducted a study on a synthetic German pig line. However, another study by Spötter et al. (2009) carried out on a much larger number of sows, representatives of the German Landrace and German Large White breeds, showed a different distribution of both alleles and genotypes in comparison with the first study. Compared with the frequency of alleles in these studies, the frequency of alleles and genotypes in sows of the German Landrace showed significant differences. Allele C occurred at a frequency of 0.61 and allele T at 0.39. Among the genotypes, CT had the highest frequency of 0.54, CC a frequency of 0.34 and the TT genotype a frequency of 0.12. The frequencies in German Large White sows were different. The C allele occurred less frequently (0.28) than the T allele (0.72). The most common genotype was TT (0.51) and the least frequent was CC (0.06). Recent research undertaken by Mucha et al. (2013) on Polish Large White and Landrace breeds showed that the CC genotype was the least frequent (0.13) and the frequency of CT (0.45) was only slightly higher than that of TT (0.42). The distribution of genotype frequencies obtained by Mucha et al. (2013) was very similar to frequencies obtained in these studies.
In Table 3 average values of the selected reproductive traits in relation to polymorphism of the LIF gene are presented for the Polish Landrace x Polish Large White sows. In the first litter, differences (P <0.01) between sows without the allele T and those with TT and CT genotypes were observed. On average the CC sows gave birth to 1.38 fewer live piglets and 1.11 more dead piglets per litter than the TT genotype. The values of individual traits between CT and TT sows were similar, and the differences were statistically insignificant. In the second litter, and after the fourth, trends were similar to that of the first litter, but the differences were not statistically significant. The analysis of the weaning-fertile mating interval does not show statistically important differences between sows with different genotypes in any of the litters.
Different results from those obtained in the present study were reported by Spötter et al. (2005). They examined sows of a synthetic German line and, despite a low C allele frequency, reported positive effects of the recessive C allele on the number of piglets born alive. Subsequent studies by Spötter et al. (2009), which included two large herds of Large White and Landrace breeds, showed that heterozygous sows from both breeds had the largest number of piglets born alive in the first litter.
However, this analysis included more litters, and reported the highest number of live piglets born to sows with genotype TT. The combined analysis of all litters of German Large White sows showed that sows without the C allele had on average 0.94 more piglets than CC sows. In the German Landrace breed, the difference was much smaller. On average, TT sows gave birth to 0.22 more piglets compared with the CC sows. Studies conducted by Mucha et al. (2013) on Polish Large White and Landrace pigs showed that sows with the TT genotype gave birth to significantly (P <0.01 and P <0.05, respectively) more live piglets in the first litter compared with sows of the CC and TC genotypes. Our results, like those of Spotter et al. (2009) and Mucha et al. (2013), indicated positive effects of the T allele on the number of live piglets per litter. However, these results should be confirmed in further studies on representative herds of sows of different breeds.
The frequency analysis showed that sows with CC and TT genotypes are more frequent than the heterozygotes. This study showed that in the first litter of sows with genotypes TT and CT in the LIF locus, the number of born and weaned piglets were higher than in sows with genotype CC. This is in accordance with the shown additive effect of the T allele on the number of piglets born alive in the first litter. The observed positive effect of the T allele indicates the possibility of using the tested polymorphism in improving the reproductive performance of sows. These results contribute to the current state of knowledge on genetic determinants of the level of reproductive traits of sows.
References
Buske, B., Sternstein, I. & Brockmann, G., 2006. QTL and candidate genes for fecundity in sows. Anim. Reprod. Sci. 95, 167-183. [ Links ]
Duncan, D.B., 1955. Multiple range and multiple F tests. Biometrics 11, 1-42. [ Links ]
Králíčková, M., Šíma, R., Vaněček, T., Šíma, P., Rokyta, Z., Ulčová-Gallová, Z., Suchá, R., Uher, P. & Hes, O., 2006. Leukemia inhibitory factor gene mutations in the population of infertile women are not restricted to nulligravid patients. Eur. J. Obstet. Gynecol. Reprod. Biol. 127, 231-235. [ Links ]
Mucha, A., Ropka-Molik, K., Piórkowska, K., Tyra, M. & Oczkowicz, M., 2013. Effect of EGF, AREG and LIF genes polymorphisms on reproductive traits in pigs. Anim. Reprod. Sci. 137, 88-92. [ Links ]
Rodriguez-Zas, S.L., Southey, B.R., Knox, R.V., Connor, J.F., Lowe, J.F. & Roskamp, B.J., 2006. Bioeconomic evaluation of sow longevity and profitability. J. Anim. Sci. 81, 2915-2922. [ Links ]
Ropka-Molik, K., Mucha, A., Oczkowicz, M. & Piórkowska, K., 2012. Determination of LIF gene expression level in ovary, oviduct and uterus of pigs in different phase of oestrus cycle. Mat. Conf., The progress of research in pigs and its use in practice" Poland, EU, 14-17.02 140-141. [ Links ]
Short, T.H., Rothschild, M.F., Southwood, O.I., McLaren, D.E., De Vries, A., Van der Steen, H., Eckardt, G.R., Tuggle, C.K., Helm, J., Vaske, D.A., Mileham, A.J. & Plastow, G.S., 1997. Effect of the estrogen receptor locus on reproduction and production traits in four commercial pig lines. J. Anim. Sci. 75, 3138-3142. [ Links ]
Spötter, A. & Distl, O., 2006. Genetic approaches to the improvement of fertility traits in the pig. J. Vet. 172, 2, 234-247. [ Links ]
Spötter, A., Drögemüller, C., Hamann, H. & Distl, O., 2001. Molecular characterization and chromosomal assignment of the porcine gene for leukemia inhibitory factor LIF. Cytogenet. Cell Genet. 93, 87-90. [ Links ]
Spötter, A., Drögemüller, C., Hamann, H. & Distl, O., 2005. Evidence of a new leukemia inhibitory factor-associated genetic marker for litter size in a synthetic pig line. J. Anim. Sci. 83, 2264-2270. [ Links ]
Spötter, A., Muller, S., Hamann, H. & Distl, O., 2009. Effect of polymorphisms in the genes for LIF and RBP4 on litter size in two German pig lines. Reprod. Dom. Anim. 44, 100-105. [ Links ]
StatSoft Inc. 2010. STATISTICA (data analysis software system) PL version 10.0. www.statsoft.com. [ Links ]
Stewart, C.L., 1994. The role of leukemia inhibitory factor (LIF) and other cytokines in regulating implantation in mammals. Ann. NY Acad. Sci. 734, 157-165. [ Links ]
Tyra, M. & Różycki, M., 2004. Heritability of reproductive traits in pigs. Anim. Sci. Pap. Rep. 22, (Suppl. 3), 235-242. [ Links ]
 Correspondence:
Correspondence:
B. Matysiak
beata.matysiak@zut.edu.pl
Received 11 September 2013
Accepted 3 February 2014
First published online 28 February 2014














