Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
South African Journal of Animal Science
versão On-line ISSN 2221-4062
versão impressa ISSN 0375-1589
S. Afr. j. anim. sci. vol.43 no.4 Pretoria 2013
Association of polymorphism in Exon 3 of toll-like receptor 4 gene with somatic cell score and milk production traits in Holstein dairy cows of Iran
R. NooriI; A.H. MahdaviI, #; M.A. EdrissI; H.R. RahmaniI; M. TalebiII; M. Soltani-GhombavaniI
IDepartment of Animal Science
IIDepartment of Biotechnology, College of Agriculture, Isfahan University of Technology, Isfahan 8415683111, Iran
ABSTRACT
Mastitis is a complex inflammatory disease of the mammary gland that is caused by the invasion of pathogens. This leads to reduced synthetic capacity, compositional changes and increased somatic cell counts (SCC) in milk. Toll-like receptor 4 (TLR4) is a cell surface receptor that recognizes lipopolysaccharides (LPS) of gram-negative bacteria. Its role in pathogen recognition and the subsequent immune response and differential expression of the gene during mastitis have prompted the investigation of TLR4 gene as a candidate to improve mastitis resistance in dairy cattle. The aim of this study was to analyse the possible association of a single nucleotide polymorphism (SNP) in putative co-receptor-binding region 2 (T4CRBR2) of the TLR4 gene with somatic cell score (SCS) and milk-related traits in 408 Iranian Holstein cows. The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique was performed for genotyping. The effect of the genotype on the traits of interest was analysed using the general linear model (GLM) procedure of SAS software. The B allele frequency was 0.634 and the distribution of genotypes was not in Hardy-Weinberg equilibrium in the overall population. The B allele of the SNP was associated with higher 305-day milk yield and breeding value for milk yield, and lower fat percentage and lower SCS, as compared with allele A. The association between TLR4 polymorphism with SCS and milk production traits reported in this study suggests that this SNP has the potential to be used as a marker for selection, based on molecular information.
Keywords: TLR4 gene, mastitis, candidate gene, milk-related traits, single nucleotide polymorphism, SNP
Introduction
Mastitis is a complex disease, which can be defined as inflammation of the mammary gland. This disease is caused by the invasion and multiplication of pathogenic microorganisms in the mammary gland, leading to reduced synthetic capacity and compositional changes (Harmon et al., 1994). This is followed by an influx of leucocytes into the milk and as a result an increase of the milk somatic cell counts (SCC) (Auldist & Hubble, 1998). Owing to high genetic correlations found between clinical mastitis and somatic cell score (SCS) (Hinrichs et al., 2005; Heringstad et al., 2006) and relatively easy recording of SCC compared with mastitis, SCC is used as an indicator of mastitis and as a tool for selection for mastitis resistance (Koivula et al., 2005). Heringstad et al. (2003) suggested that mastitis resistance has a significant genetic component, and it is possible to gain noticeable selection response for clinical mastitis, and that the unfavourable genetic correlation between mastitis and milk production traits would lead to a further increase in mastitis occurrence if mastitis is ignored in breeding programmes.
The innate immune response is the first line of defence against microbial pathogens (Janeway & Medzhitov, 2002). Genes encoding immune factors that detect and eliminate pathogens are strong potential candidates for enhancing resistance against infectious diseases like mastitis. Toll-like receptor 4 (TLR4) is a cell surface receptor that recognizes lipopolysaccharides (LPS) of gram-negative bacteria (Akira & Takeda, 2004). Gene expression patterns indicate the innate immune response of the mammary gland, which is characterized by production of a large amount of important mediators of innate immunity following activation of the TLR4. Tolllike receptor 4 has evolved with its accessory proteins (LBP, CD14, and MD-2/Ly96) to recognize lipopolysaccharides (LPS). The detection of the LPS molecule by TLR4 leads to the activation of intracellular signalling molecules (Zheng et al., 2006). Mastitis significantly enhances the expression of the TLR4 gene (Goldammer et al., 2004).
Based on the above studies on the role of TLR4 in pathogen recognition and subsequent initiation of the inflammatory and immune response, and on differential expression of the gene during mastitis, TLR4 has been proposed as a strong candidate for increasing mastitis resistance in breeding programmes (Ogorevc et al., 2009).
Several polymorphisms have been identified in bovine TLR4 gene (White al., 2003), some of which are reported to affect SCS. One single nucleotide polymorphism (SNP) in a putative promoter region (P-226) and two SNPs in exon 3 (E3+1656 and E3+2021) of TLR4 have been found to be associated with estimated breeding values for lactation persistency, and SCS in the Canadian Holstein bull population (Sharma et al., 2006). In addition, Wang et al. (2007) found an association between polymorphism in putative co-receptor-binding region 1 (T4CRBR1) of the gene and SCS in Chinese Holstein, Sanhe and Chinese Simmental cattle. No significant association was revealed between the genotypes of locus T4CRBR2 and SCS for these cattle. Another SNP (g.8664C>T) in the toll IL-1 receptor (TIR) domain of the bovine TLR4 was shown to be associated with SCS (Wang et al., 2008). Beecher et al. (2010) also revealed an association of E3+2021 with milk fat and protein percentage in late lactation in Holstein-Friesian, Jersey, Norwegian Red, Montbeliarde, and Holstein-Friesian x Jersey cows, but not in Holstein-Friesian bulls.
Owing to the effect of mastitis on milk yield and composition (Shuster et al., 1991; Auldist & Hubble, 1998; Leitner et al., 2004) and the existence of a genetic correlation between mastitis and milk yield and composition (Hinrichs et al, 2005), it is possible that TLR4, as a candidate gene influencing mastitis resistance, also affects milk production and components. Literature that addresses the association of this gene with milk-related traits is limited (Beecher et al., 2010). The present study was designed to investigate the possible association of SNP at the 1397-bp position in putative co-receptor-binding region 2 (T4CRBR2) of the TLR4 gene with SCS and milk-related traits in Iranian Holstein cows.
Materials and Methods
Blood samples were collected from 408 Iranian Holstein cows randomly selected from five dairy herds in Isfahan province, using vacuum venoject tubes containing EDTA, and stored at -20 °C for DNA extraction. Accurate phenotypic data for 305-day milk yield, milk fat and protein percentage, SCS, and breeding value data for fat and milk yield were obtained from the Vahdat Industrial Agriculturists & Dairymen Cooperative of Isfahan.
Genomic DNA was extracted from blood samples by the salting out method (Miller et al, 1988). Genotypes were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique. PCR was used to amplify the 382-bp DNA fragments containing the polymorphism of interest. Each PCR reaction was carried out in a total volume of 20 uL containing 50 ng genomic DNA, 10 pmoL each primer, 2 uL 10X PCR buffer, 2 mM MgCl2, 200 uMdNTPs and 2.5 units Taq DNA polymerase. Sequences of the primers used in the PCR were reported previously by Wang et al. (2007). The sequences of the forward and reverse primers were 5' AGACAGCATTTCACTCCCTC 3' and 5' ACCACCGACACACTGATGAT 3', respectively. PCR cycling conditions were as follows: initial denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 30 s, 59 °C annealing temperature for 30 s, extension at 72 °C for 40 s, and a final extension at 72 °C for 10 min. The PCR products were digested with 5 U of AluI enzyme (Fermentas, St. Leon-Rot, Germany) in 20 uL of reaction volume at 37 °C for 10 h. The digested products were separated by horizontal electrophoresis on 2.5% agarose gel in 0.5 x TBE (90 v for 1.5 h) stained with ethidium bromide and visualized under UV light. The A allele was indicated by two bands at 300 bp and 82 bp and the B allele resulted in three fragments of 160, 140 and 82 bp.
The allele and genotype frequencies were estimated by direct counting, and deviation from Hardy-Weinberg equilibrium was examined using the POPGENE v. 1.31 (Yeh et al, 1997). For the association studies, the traits of interest were analysed using the least square method of the general linear model (GLM) procedure of the SAS software (SAS, 1994) according to the following model:
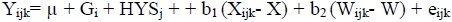
where Yijk is value for each trait, j is overall mean, Gi is the effect of the ith genotype (AA, AB and BB), HYSj is the combined effect of herd (1, 2, 3, 4, and 5), year and season of parturition, b1 is the linear regression coefficient of milk yield, Xijkl is milk yield, X is mean milk yield, b2 is the linear regression coefficient of open days, Wijkl is open days, W is mean open days and eijk is the random residual effect.
For the analysis of 305-day milk yield, milk yield as a covariate was omitted from the model. Also, wherever breeding value was used instead of phenotypic value for a trait, effects other than genotype were excluded from the model.
Results
Both alleles (A and B) and three genotypes (AA, AB and BB) of the T4CRBR2 locus were observed in all five herds. The genotype and allele frequencies of T4CRBR2 in Iranian Holstein cows are listed in Table 1. The result of the test for Hardy-Weinberg equilibrium for this locus indicated that the genotypes in herds three, four and five did not show any significant deviation from Hardy-Weinberg equilibrium, while herds one, two and the overall population were not distributed according to Hardy-Weinberg equilibrium (Table 1).
Least square means of the three TLR4 genotypes for the traits of interest are presented in Table 2. Genotypes of T4CRBR2 were significantly associated with 305-day milk yield, breeding value for milk yield, fat percentage and SCS (P <0.05). Genotype AA had the lowest and BB the highest 305-day milk yield, while AB was intermediate (P <0.05). Furthermore, cows with genotype AA had significantly lower breeding value for milk yield compared to AB and BB genotypes (P <0.05).
Cows homozygous for the A allele had significantly higher milk fat percentage than those of AB genotype (P <0.05) and cows of AB genotype had a higher fat percentage than those homozygous for the B allele (P <0.05). Animals carrying genotype BB showed lower SCS in comparison with genotypes AB and AA (P <0.05). No significant association was evident between the genotypes of the SNP and protein percentage and breeding value for fat yield in our study (P >0.05).
Discussion
The allele frequencies obtained in the present study were in agreement with the findings of Wang et al. (2007), who reported frequencies of 0.42, 0.44 and 0.45 for allele A and 0.58, 0.56 and 0.55 for allele B in Chinese Holstein, Sanhe and Chinese Simmental populations, respectively.
In our study, the B allele of T4CRBR2 SNP was found to be associated with lower SCS. However, Wang et al. (2007) did not show any relationship between this polymorphism and SCS. Since the SNP in the T4CRBR2 locus is a synonymous one in coding region of the gene, it will not change the amino acid sequence and the function of TLR4. Thus, the SNP is not a functional mutation, but may be in linkage disequilibrium with a functional polymorphic site in TLR4 or other closely linked gene(s) influencing this trait. TLR4 has been mapped to the distal end of bovine chromosome 8 (White et al., 2003) and Quantitative Trail Loci (QTL) affecting SCS, and clinical mastitis have been found on this chromosome. Klungland et al. (2001) mapped a single QTL affecting SCC at position 54 cM, and a QTL for clinical mastitis at position 46 cM in a Norwegian sire family.
The allele B of the SNP was associated with higher 305-day milk yield and breeding value for milk yield. The association of the B allele with higher 305-day milk yield and breeding value for milk yield observed in the current study may be attributed to the negative effect that increased SCS and mastitis have on milk production. Although QTL for milk yield have not been detected on BTA8 in the vicinity of TLR4, unfavourable genetic correlation has been found between mastitis and milk yield (Hinrichs et al., 2005). Clinical mastitis can induce mammary cell death and affect milk secretion detrimentally (Capuco et al., 1986; Long et al., 2001).
In the present study, the allele B of the SNP was associated with lower fat percentage. However, no association was found between the polymorphism and protein percentage and breeding value for fat yield. A study by Beecher et al. (2010) indicated an association of E3+2021 polymorphism in TLR4 gene with milk fat and protein percentage in late lactation in Holstein-Friesian, Jersey, Norwegian Red, Montbeliarde, and Holstein-Friesian x Jersey cows, but not in Holstein-Friesian bulls. The association of the TLR4 polymorphism with fat percentage in our study can simply be attributed to negative genetic correlation between fat percentage and milk yield (Chauhan & Hayes, 1991; Rosati & Van Vleck, 2002). Our observation that the B allele was associated with higher milk yield and lower fat percentage supports this hypothesis. On the other hand, our association of allele A with higher SCS and higher fat percentage is in agreement with findings of other studies that have reported an increase in fat content as a result of mastitis (Shuster et al., 1991; Pyorala et al., 2003; Bruckmaier et al., 2004).
Conclusions
An association of T4CRBR2 polymorphism in the TLR4 gene with SCS, 305-day milk yield, breeding value for milk yield and fat percentage was evident in this study. Our results suggest that this SNP may be of use as a potential genetic marker in selection programmes for these traits in dairy cattle. Further association studies of various polymorphisms located in different regions of the TLR4 with these traits in other breeds and populations of cattle are needed in order to verify the obtained results.
Acknowledgment
The authors gratefully acknowledge Isfahan University of Technology for supporting this research financially. The authors also acknowledge the Vahdat Industrial Agriculturists & Dairymen Cooperative of Isfahan province of Iran for supplying data for milk-related traits.
References
Akira, S. & Takeda, K., 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499-511. [ Links ]
Auldist, M.J. & Hubble, I.B., 1998. Effects of mastitis on raw milk and dairy products. Aust. J. Dairy Technol. 53, 28-36. [ Links ]
Beecher, C., Daly, M., Childs, S., Berry, D.P., Magee, D.A., McCarthy, T.V. & Giblin, L., 2010. Polymorphisms in bovine immune genes and their associations with somatic cell count and milk production in dairy cattle. BMC Genet. 11, 99. [ Links ]
Bruckmaier, R.M., Ontsouka, C.E. & Blum, J.W., 2004. Fractionized milk composition in dairy cows with subclinical mastitis. Vet. Med-Czech. 8, 283-290. [ Links ]
Capuco, A.V., Paape, M.J. & Nickerson, S.C., 1986. In vitro study of polymorphonuclear leukocyte damage to mammary tissues of lactating cows. Am. J. Vet. Res. 47, 663-668. [ Links ]
Chauhan, V.P. & Hayes, J.F., 1991. Genetic parameters for first milk production and composition traits for Holsteins using multivariate restricted maximum likelihood. J. Dairy Sci. 74, 603-610. [ Links ]
Goldammer, T., Zerbe, H., Molenaar, A., Schuberth, H.J., Brunner, R.M., Kata, S.R. & Seyfert, H.M., 2004. Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin. Diagn. Lab. Immunol. 11, 174-185. [ Links ]
Harmon, R.J., 1994. Physiology of mastitis and factors affecting somatic cell counts. J. Dairy Sci. 77, 2103-2112. [ Links ]
Heringstad, B., Klemetsdal, G. & Steine, T., 2003. Selection responses for clinical mastitis and protein yield in two Norwegian dairy cattle selection experiments. J. Dairy Sci. 86, 2990-2999. [ Links ]
Heringstad, B., Gianola, D., Chang, Y.M., 0degard, J. & Klemetsdal, G., 2006. Genetic associations between clinical mastitis and somatic cell score in early first-lactation cows. J. Dairy Sci. 89, 2236-2244. [ Links ]
Hinrichs, D., Stamer, E., Junge, W. & Kalm, E., 2005. Genetic analyses of mastitis data using animal threshold models and genetic correlation with production traits. J. Dairy Sci. 88, 2260-2268. [ Links ]
Janeway, C.A.J. & Medzhitov, R., 2002. Innate immune recognition. Annu. Rev. Immunol. 20, 197-216. [ Links ]
Klungland, H., Sabry, A., Heringstad, B., Olsen, H.G., Gomez-Raya, L., Vage, D.I., Olsaker, I., Odegard, J., Klemetsdal, G., Schulman, N., Vilkki, J., Ruane, J., Aasland, M., Ronningen, K. & Lien, S., 2001. Quantitative trait loci affecting clinical mastitis and somatic cell count in dairy cattle. Mamm. Genome 12, 837-842. [ Links ]
Koivula, M., Mantysaari, E.A., Negussie, E. & Serenius, T., 2005. Genetic and phenotypic relationship among milk yield and somatic cell count before and after clinical mastitis. J. Dairy Sci. 88, 827-833. [ Links ]
Leitner, G., Chaffer, M., Shamay, A., Shapiro, F., Merin, U., Ezra, E., Saran, A. & Silanikove, N., 2004. Changes in milk composition as affected by subclinical mastitis in sheep. J. Dairy Sci. 87, 46-52. [ Links ]
Long, E., Capuco, A.V., Wood, D.L., Sonstegard, T., Tomita, G., Paape, M.J. & Zhao, X., 2001. Escherichia coli induces apoptosis and proliferation of mammary cells. Cell Death Differ. 8, 808-816. [ Links ]
Miller, S.A., Dykes, D.D. & Polesky, H.F., 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215. [ Links ]
Ogorevc, J., Kunej, T., Razpet, A. & Dovc, P., 2009. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim. Genet. 40, 832-851. [ Links ]
Pyorala, S., 2003. Indicators of inflammation in the diagnosis of mastitis. Vet. Res. 34, 565-578. [ Links ]
Rosati, A. & Van Vleck, L.D., 2002. Estimation of genetic parameters for milk, fat, protein and mozzarella cheese production for the Italian river buffalo Bubalus bubalis population. Livest. Prod. Sci. 74, 185-190. [ Links ]
SAS, 1994. Statistical Analysis Systems User's Guide. Statistics Version 6. SAS Institute, Inc. Cary, N.C., USA. [ Links ]
Sharma, B.S., Leyva, I., Schenkel, F. & Karrow, N.A., 2006. Association of toll-Like receptor 4 polymorphisms with somatic cell score and lactation persistency in Holstein bulls. J. Dairy Sci. 89, 3626-3635. [ Links ]
Shuster, D.E., Harmon, R.J., Jackson, J.A. & Hemken, R.W., 1991. Suppression of milk production during endotoxin-induced mastitis. J. Dairy Sci. 74, 3763-3774. [ Links ]
Wang, X., Xu, S., Gao, X., Ren, H. & Chen, J., 2007. Genetic polymorphism of TLR4 gene and correlation with mastitis in cattle. J. Genet. Genomics 34, 406-412. [ Links ]
Wang, X., Xu, S., Gao, X., Li, J. & Ren, H., 2008. Cloning and SNP screening of the TLR4 gene and the association between its polymorphism and somatic cell score in dairy cattle. S. Afr. J. Anim. Sci. 38, 101-109. [ Links ]
White, S.N., Taylor, K.H., Abbey, C.A., Gill, C.A. & Womack, J.E., 2003. Haplotype variation in bovine toll-like receptor 4 and computational prediction of a positively selected ligand-binding domain. Proc. Natl. Acad. Sci., USA. 100, 10364-10369. [ Links ]
Yeh, F.C., Yang, R.C., Boyle, T.B.J., Ye, Z.H. & Mao, J.X., 1997. Popgene, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Canada. [ Links ]
Zheng, J., Watson, A.D. & Kerr, D.E., 2006. Genome-wide expression analysis of lipopolysaccharide-induced mastitis in a mouse model. Infect. Immun. 74, 1907-1915. [ Links ]
Received 28 June 2013
Accepted 10 October 2013
First published online 21 December 2013
# Corresponding author: mahdavi@cc.iut.ac.ir














