Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.43 n.1 Pretoria Jan. 2013
SHORT COMMUNICATION
Analysis of inbreeding of the South African Dairy Swiss breed
P. de Ponte BouwerI; C. VisserI, #; B.E. MostertII
IDepartment of Animal and Wildlife Sciences, University of Pretoria, Private Bag x 20, Hatfield, 0028, South Africa
IISA Studbook, PO Box 270, Bloemfontein, 9300, South Africa
ABSTRACT
In South Africa, the Dairy Swiss breed, which originated in Switzerland, consists of 27 breeders and 1135 breeding cows. Pedigree information on the breed was analysed to determine its effective population size (Ne) and rate of inbreeding. The rate of inbreeding was 0.08% per year and 0.38% per generation. It was estimated that in 2008, over 90% of the animals were inbred. This is within the acceptable recommendation for an effective population size of 50 - 100, with an Ne value of 89. However, the high percentage of inbred animals may start to have a detrimental impact on the effective population size and breeders need to exercise caution in their breeding decisions.
Keywords: Effective population size, genetic diversity, inbreeding coefficient
Brown Swiss was developed in north-eastern Switzerland, and is probably the oldest of all dairy breeds (http://www.studbook.co.za/Society/swiss/history.php). The first Brown Swiss bull was imported into South Africa from the USA in 1907, and was closely followed by another bull and five heifers from Switzerland. These imports laid the foundation for the first Brown Swiss herd in South Africa (Smit, 2008). The Brown Swiss Cattle Breeders' Society of South Africa was founded in 1925.
Brown Swiss originated as a dual-purpose breed. However, in most parts of the world two breeds have developed from the original. In 1974, it was decided to create separate South African herd books for the dairy (South African Dairy Swiss) and beef (Braunvieh South Africa) breeds. The South African Dairy Swiss (SA Dairy Swiss) was recognised as a separate breed by the Livestock Improvement Act of the National Department of Agriculture in 1995 (Smit, 2008). The SA Dairy Swiss is a small breed with only 27 active breeders in South Africa. The active national herd in 2009 consisted of 127 fully registered bulls, and 1 135 cows of which 755 were fully registered (2009, C. Hunlun, pers. comm., charl@studbook.co.za).
Technologies for assisted reproduction have been applied in dairy breeding, and have been effective in generating genetic gain (Sorensen et al., 2005). However, these reproductive technologies and the accompanying greater selection intensity have increased inbreeding (Weigel, 2001; Kearney et al., 2004; Ponzoni et al., 2010). A consequence of inbreeding is a reduction of genetic variability within the population and reduced performance of animals, mainly in traits associated with fitness (Wall et al., 2005), and also adversely affecting production traits (Koenig & Simianer, 2006; Maiwashe et al., 2008). This leads to reduced profitability (Weigel & Lin, 2002) and unsustainable genetic improvement in the long term (Sorensen et al., 2005, Maiwashe et al., 2006; Ponzoni et al., 2010). While the impact of inbreeding in populations of large sizes may be negligible, its effect in typical livestock populations in which selective breeding is practised can be more severe.
Effective population size (Ne) is a measure of the genetic diversity within a population (Wright, 1931) and can be calculated from the number of parents or the rate of inbreeding (Falconer & Mackay, 1996).
According to Boichard et al. (1997), Ne is a powerful tool for predicting the change in genetic variability over a long period, when the inbreeding increase fully reflects the number and choice of breeding animals in the previous generations. In animal breeding, the recommendation is to maintain an Ne of at least 50 to 100 (Meuwissen, 1999; Sorensen et al., 2005). Therefore, monitoring effective population size (Ne) and possible control of resultant inbreeding may be crucial to implementing genetic improvement programmes (Ponzoni et al, 2010). The SA Dairy Swiss had not been analysed in the past for its effective population size or its inbreeding status. The aim of this study is therefore to give an overview of its effective population size and inbreeding status.
Pedigree data were downloaded from the INTERGIS (Integrated Registration and Genetic Information System of South Africa) for the SA Dairy Swiss breed. Over the eighty-one years from 1928 to 2009 the SA Dairy Swiss pedigree database comprised 38 024 animals: 26 731 cows and 11 293 bulls. As pedigree information of foreign animals is rather incomplete on the INTERGIS, the INTERBULL database as well as the Canadian Dairy Network (www.cdn.ca) was used to retrieve and validate pedigree information of imported animals. All pedigree information was included in the analysis. However, since the SA Dairy Swiss received its own herd book in 1974, only the results from 1974 to 2008 are presented. The base population, which is defined as all animals with unknown parents, consisted of 7 622 animals.
Editing the data included checking the integrity of the pedigree for completeness and accuracy. Duplicated records of foreign animals were eliminated, and unknown parents of foreign animals in the pedigree database were added. Because the pedigree information of the imported animals was incomplete, tracing them back resulted in more records in the final dataset than the SA Dairy Swiss pedigree database. Thus, a total of 40 141 records were available for analyses. Inbreeding coefficients for all animals of the population were calculated with Poprep (www.popreport.tzv.fal.de) (Groeneveld et al., 2009). An inbred animal was defined as any animal with an inbreeding coefficient greater than zero. The level of inbreeding of the population per year was calculated by averaging inbreeding coefficients by year of birth.
The rate of inbreeding per generation (AF) was calculated as follows (Falconer & Mackay, 1996):

AF = (Ft - Ft-1) / (1 - Ft-1) [1] where Ft and Ft-1 are the average inbreeding of offspring and their parents, respectively. The effective population size (Ne) was calculated according to Falconer & Mackay (1996):

An estimate of an individual's inbreeding coefficient depends on the extent to which its ancestry is known. The more complete the knowledge of an individual's ancestry is, the more reliable its estimate of the inbreeding coefficient relative to the defined base population (Boichard et al., 1997; VanRaden, 2005; Groeneveld et al., 2009, Van der Westhuizen, 2009). The number of animals with unknown parents per year (thus base animals), is depicted in Figure 1.
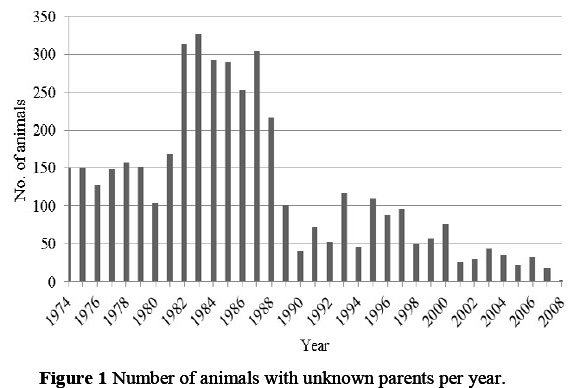
In terms of animals born since 1974, the number of generations of complete pedigree information for the SA Dairy Swiss breed, ranged from one to six. This compares favourably with the study by Weigel (2001), in which a depth of six generations of pedigree completeness was reported. Since the number of animals was increasing, the expansion in pedigree completeness up to 1994 was a good indication of breeder participation and competent record keeping.
In 1994 the pedigree for generation 1 was 94.1% complete, while that of generation 3 was 85.3% complete, and generation 6 was 63.6% complete (Figure 2) (a generation interval equals five years). The greater percentage of pedigree completeness for generation 1 is as expected, since it is easier to trace the pedigree information back to one, two and sometimes even three generations. However, tracing a pedigree for more generations is harder, as information may have been misplaced or not recorded.
The reduced percentage of pedigree completeness from 1994 to 2004 could be the result of administrative obligations and costs associated with recording pedigrees and performance testing, because milk recording was no longer fully subsidized by the government (Liebenberg, 2010).
The rate of inbreeding of 0.08% per year (Figure 3) and 0.38% per generation for the SA Dairy Swiss Breed is well within the limits of acceptability of up to 0.5% per year and between 1% and 1.5% per generation (Van der Westhuizen & Mostert, 1998; Weigel, 2001). As reported by Nicholas (1989), rates of inbreeding of this magnitude would lead to a coefficient of variation of selection response of less than 10% over a 10-year period. However, when compared with previously published results of other South African breeds, the rate of inbreeding per year in the SA Dairy Swiss breed is greatest, followed by Jerseys (0.07%), Holsteins (0.06%), Ayrshires and Guernseys (0.05%) (Maiwashe et al., 2006). These estimates, however, may not be directly comparable with the current results, as different depths of pedigree and pedigree completeness levels might influence results.
In Figure 4, the total number of animals in the SA Dairy Swiss population is indicated, as well as the total number and percentage of inbred animals since 1975. Since 1975, 33.8% of animals born in South Africa have been inbred. As the total number of animals increased, so did the number of inbred animals. After 1994, the total number of animals and the number of inbred animals decreased, while the percentage of inbred animals in the population increased. The lowest percentage of inbred animals was 5.2% in 1975. The percentage of inbred animals increased steadily until 2001 - 2002. In 2002, 51.4% of SA Dairy Swiss was inbred compared with 66% inbred animals in the Ayrshire breed (Mostert, 2003) and 63% in the Jersey breed (Mostert, 2011). After 2002, the percentage of inbred SA Dairy Swiss animals increased sharply in 2005 to 67.3% and then reached 90.2% in 2008 (with no animals having an inbreeding coefficient >6.25%). This is much greater than the 71% of inbred animals born in the Jersey population in 2009. This indicates that since the SA Dairy Swiss breed is small, breeders have a limited selection of animals to mate, compared with the other dairy breeds. Breeders are probably mating related animals with no knowledge of the consequences or structured plan to restrict the rate of inbreeding.
The distribution of inbreeding coefficients and the number of animals that fall into each category of inbreeding coefficients in the SA Dairy Swiss population are summarised in Figure 5. The distribution ranged between 22.4% of the population for 0<F<5% to 0.17% for F>30%, with 66.3% of the population having inbreeding coefficients of 0. The animals that have not been inbred include those with unknown parents, as it is assumed that unknown parents are unrelated. As a rule of thumb, breeders are advised to avoid matings that result in inbreeding coefficients of offspring greater than 6.25% (Van der Westhuizen & Mostert, 1998). The highest inbreeding coefficient for an AI bull used in South Africa in the Brown Swiss breed is Avon View Ideal from the USA, with F = 25%. The highest inbreeding coefficient for an individual animal in the SA Dairy Swiss breed is a cow with an inbreeding coefficient of 39.2%, born in 2007.
When Ne is calculated, using the difference between the average inbreeding of offspring born per year and the average inbreeding of their parents (Falconer & Mackay, 1996), the SA Dairy Swiss breed had an Ne = 56 in 2001 in comparison with the estimates of Ne found by Weigel (2001) of 61 for the Brown Swiss and 161, 65, 39 and 30 for the Ayrshire, Guernsey, Holstein and Jersey breeds, respectively. The estimates of effective population sizes reported by Maiwashe et al. (2006) were 148, 165, 137, and 108 for the SA Ayrshire, Guernsey, Holstein and Jersey breeds, respectively.
The SA Dairy Swiss breed had a low effective population size of 46 and 45, based on offspring born in 2006 and 2007, respectively. However, the breed's Ne improved for offspring born in 2008, increasing up to 89. If Ne is based on the change of inbreeding per generation since 1974 (Δ F = 0.38%) (Figure 3), Ne = 132, which is comparable with the Ne of other dairy populations reported by Maiwashe et al. (2006).
The larger dairy breeds (Holstein and Jersey) have lower effective population sizes in comparison with the smaller breeds (Ayrshire and Guernsey) (Maiwashe et al., 2006). According to Weigel (2001) and Cleveland et al. (2005), the lower Ne of major dairy breeds is the result of elite sires being used extensively across countries through artificial insemination (AI). An Ne of fewer than 100 animals indicates a potential limitation in genetic diversity. This, together with the fact that the total population and herd size of the SA Dairy Swiss breed are small, implies that within-breed genetic diversity could be adversely affected and random gene frequency changes that are cumulative over generations should be expected (Maiwashe & Blackburn, 2004).
Mating decisions will play an important short-term role in curbing the levels of inbreeding in the SA Dairy Swiss population. Breeders should note the importance of decreasing rates of co-ancestry, possibly by limiting the use of popular sires, as has been done in the Canadian Holstein population (Stachowicz et al., 2011). A minimum co-ancestry mating system, combined with the avoidance of sib-matings, should result in acceptable levels of inbreeding. Koenig & Simianer (2006) proposed that a combination of the optimum genetic contribution theory and specific mating plans is a viable option to control relationships and inbreeding. Furthermore, it is the responsibility of all breeders to participate in pedigree recording in order to limit the levels of inbreeding and ensure that a sustainable amount of genetic variability remains in the population. To preserve the genetic composition of dairy cattle for the future, a reserve of cryopreserved germplasm could be developed that could be used to regenerate the breed, should the need arise. Molecular genetic tools could also be used to assess genetic diversity from time to time (Maiwashe & Blackburn, 2004).
Although the rate of inbreeding in the SA Dairy Swiss population is acceptable, the percentage of animals being inbred is a major cause for concern, with 90% of the animals born in 2008 being inbred. One way to manage inbreeding will be through using mate-allocation programmes that constrain the level of inbreeding of future progeny. Estimates of effective population sizes indicate that genetic variability in South African dairy breeds is still greater than that of their counterparts in the US. While the rate of inbreeding and effective population sizes have not reached critical levels, the existence of highly inbred individuals in this population necessitates further study to quantify the phenotypic effect of inbreeding on traits of economic importance in this population.
References
Boichard, D., Maignel, L. & Verrier, E., 1997. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 29, 5-23. [ Links ]
Cleveland, M.A., Blackburn, H.A., Enns, R.M. & Garrick, D.J., 2005. Changes in inbreeding of U.S. Herefords during the twentieth century. J. Anim. Sci. 83, 992-1001. [ Links ]
Falconer, D.S. & Mackay, T.F.C., 1996. Introduction to Quantitative Genetics. Longman Group, Essex, UK. [ Links ]
Groeneveld, E., Van der Westhuizen, B., Maiwashe, A., Voordewind, F. & Ferraz, J.B.S., 2009. POPREP: A generic report for population management. Genet. Mol. Res. 8 (3), 1158-1178. [ Links ]
Kearney, J.F., Wall, E., Villanueva, B. & Coffey, M.P., 2004. Inbreeding trends and application of optimized selection in the UK Holstein population. J. Dairy Sci. 87, 3503-3509. [ Links ]
Koenig, S. & Simianer, H., 2006. Approaches to the management of inbreeding and relationship in the German Holstein dairy cattle population. Livest. Sci. 103, 40-53. [ Links ]
Liebenberg, F., 2010. South African Agricultural Production and Investment Patterns. Farm Foundation-Economic Research Service. Conference on Global Agricultural Productivity, May 11-12, 2010; Waugh Conference Centre, ERS, Washington D.C., USA. [ Links ]
Maiwashe, M.A. & Blackburn, H.D., 2004.Genetic diversity in and conservation strategy considerations for Navajo Churro sheep. J. Anim. Sci. 82, 2900-2905. [ Links ]
Maiwashe, A., Nephawe, K.A., Van der Westhuizen, R.R., Mostert, B.E. & Theron, H.E., 2006. Rate of inbreeding and effective population size in four major South African dairy cattle breeds. S. Afr. J. Anim. Sci. 36, 50-57. [ Links ]
Maiwashe, A., Nephawe, K.A. & Theron, H.E., 2008. Estimates of genetic parameters and effects of inbreeding on milk yield and composition in South African Jersey cows. S. Afr. J. Anim. Sci. 38, 119-125. [ Links ]
Meuwissen, T.H.E., 1999. Operation of conservation schemes, in Genebanks and the Conservation of Farm Animal Genetic Resources. Ed. Oldenbroek, K.K., DLO Institute for Animal Science and Health, Lelystad, The Netherlands. pp. 9-112. [ Links ]
Mostert, B.E., 2003. Inbreeding in the South African Ayrshire breed. The Dairy Mail 11 (6), 138-139. [ Links ]
Mostert, B.E., 2011. Facts on the inbreeding of the South African Jersey breed. Jersey Journal. [ Links ]
Nicholas, F.W., 1989. Evolution and Animal Breeding. Eds Hill, W.G. & Mackay, T.F.C., CAB Intl., Wallingford, UK. pp. 201-209. [ Links ]
Ponzoni, R.W., Khaw, H.L., Nguyen, N.H. & Hamzah, A., 2010. Inbreeding and effective population size in the Malaysian nucleus of the GIFT strain of Nile tilapia (Oreochromis niloticus). J. Aquacult. 302, 42-48. [ Links ]
Smit, A., 2008. Aspoester word 'n Prinses. The Dairy Mail (August). Volume 15, N° 8. [ Links ]
Sorensen, A.C., Sorensen, M.K. & Berg, P., 2005. Inbreeding in Danish Dairy cattle breeds. J. Dairy Sci. 88, 1865-1872. [ Links ]
Stachowicz, K., Sargolzaei, M., Miglior, F. & Schenkel, F.S., 2011. Rates of inbreeding and genetic diversity in Canadian Holstein and Jersey cattle. J. Dairy Sci. 94, 5160-5175. [ Links ]
Van der Westhuizen, B., 2009. Population history & genetic variability in Bonsmara Beef Cattle via a pedigree analysis. Bonsmara: Born to Breed - Born to Lead. Edited & Compiled by P. Ferreira. [ Links ]
Van der Westhuizen, J. & Mostert, B.E., 1998. Inbreeding and the Stud Breeder. Charolais Journal. [ Links ]
VanRaden, P.M., 2005. Inbreeding adjustments and effect on genetic trend estimates. Proceedings of the 2005 INTERBULL meeting, Uppsala, Sweden. June 2-4. Bulletin No. 33. [ Links ]
Wall, E., Brotherstone, S., Kearney, J.F., Woolliams, J.A. & Coffey, M.P., 2005. Impact of non-additive genetic effects in the estimation of breeding values for fertility and correlated traits. J. Dairy Sci. 88, 376-385. [ Links ]
Weigel, K.A., 2001. Controlling inbreeding in modern breeding programs. J. Dairy Sci. 84, 177-184. [ Links ]
Weigel, K.A. & Lin, S.W., 2002. Controlling inbreeding by constraining the average relationship between parents of young bulls entering AI progeny test programs. J. Dairy Sci. 85, 2376-2383. [ Links ]
Wright, S., 1931. Evolution in Mendelian populations. Genetics 16, 97-159. [ Links ]
Received 10 August 2011
Accepted 18 January 2013
First published online 9 March 2013
# Corresponding author: carina.visser@up.ac.za














