Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Animal Science
versión On-line ISSN 2221-4062
versión impresa ISSN 0375-1589
S. Afr. j. anim. sci. vol.42 no.5 Pretoria ene. 2012
Genetic evaluation of growth traits in beef cattle using random regression models
F.W.C. NeserI, #; J.B. van WykI; M.D. FairI; P. LuboutII
IDepartment of Animal Wildlife and Grassland Sciences, UFS, Bloemfontein 9300, South Africa
IIBrangus Cattle Breeders' Society, PO Box12465, Brandhof, Bloemfontein, 9324, South Africa
ABSTRACT
Direct- and maternal heritabilities were estimated for weight traits in Brangus cattle using random regression models. After editing, 54 924 records, from birth- (BW) to mature weight (MW) from 21 673 animals were selected for analysis. The data, which covered a period of 8 generations (1985 to 2010), were transformed to a log scale to accommodate the wide range of weights being studied (15 to 850 kg). Traits included in the analysis were birth- (BW), weaning- (WW), yearling- (YW), eighteen month- (FW) and three measurements of mature weight (MW). Linear polynomials with intercepts were fitted using random regression models. The direct heritability estimates were moderate and ranged from 0.13 to 0.25 while maternal heritability estimates were low ranging from 0.05 to 0.06.
Keywords: Heritability estimates, South African Brangus cattle
Introduction
Random regression models (RRM) have become the method of choice to analyze longitudinal data or repeated measurements (Meyer, 2004; 2005; Schaeffer & Jamrozik, 2008). Although this technology has been used in dairy production studies for the genetic analysis of test day models (Legarra et al., 2004), its application for growth traits in beef cattle is obvious as an animal is weighed several times during its lifetime, sometimes repeatedly for the same trait. Not only would it be possible to utilize all these measurements to obtain accurate breeding values, but selection using the animal's actual growth curve becomes a possibility. Meyer (2004) found that RRM are up to 5.9% more accurate than multi-trait models (MT). However, Bohmanova et al. (2005) stated that the superiority of RRM over MT depends on the quality of implementation. Nobre et al. (2003) found that even after reparameterization by diagonalization, evaluations with RRM using Legendry polynomials were less accurate than MT. Misztal et al. (2000) found that parameters estimated with RRM using Legendry polynomials are prone to contain artifacts due to data distribution. This is especially true where estimates of direct and maternal covariances are obtained. Legarra et al. (2004) developed a method that ensures artifact-free parameters from RRM using Legendry polynomials. An alternative to Legendry polynomials are splines. Meyer (2005) found that due to a locality of fit for each parameter, models using splines have potentially better numerical properties than polynomials. Bohmanova et al. (2005) reported no difference in the accuracies of RRM using Legendry polynomials, RRM using splines and MT.
The aim of this study was to estimate variance components for growth traits (birth- to mature weight) for South African Brangus cattle using random regression models.
Materials and Methods
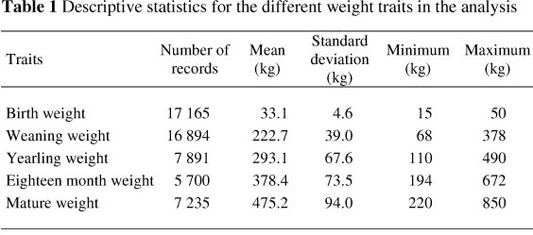
Data from 73 676 animals for birth weight (BW - 41 572), weaning weight (WW - 23 104), yearling weight (YW - 9 114), eighteen month weight (FW - 6 450) and mature weight (MW - 9 258) were available to estimate (co)variance components and subsequent genetic parameters in the breed. All incomplete records, as well as records outside four standard deviations from the mean, were disregarded. Herds with less than three years of recording, as well as contemporary groups with less than five records and animals with less than two measurements, were also removed from the final data set used for the analysis. The rules for the performance recording of Brangus cattle in South Africa allows for two measurements per trait (WW, YW and FW). Only the weight closest to the normal recording age for the trait was considered. Only the first three measurements were used for mature weight.
The final dataset consisted of 54 924 weight records, obtained from 21 673 animals in 78 herds, collected over a period of 26 years (1985 - 2010). The data were transformed to a log scale to accommodate the wide range of weights being studied (15 to 850 kg). A total of 1 623 sires and 415 sires of sires, as well as 18 491 dams with 1 028 sires of dams and 5 729 dams of dams, were present in the data. The pedigree file consists of 58 973 animals born over a period of eight generations.
The following fixed effects were included in the model: A concatenation of herd-year-season, sex and dam-age. Two distinct calving seasons were identified: the months from September to March were classified as season one, while April to August was classified as season two. Age of dam was expressed in years, starting with dams of two years and younger. All dams older than six years were grouped together. The following traits were pre-corrected for weighing age to simplify the analysis: weaning-, yearling- and eighteen month weight.
The weight traits were analysed in a single-trait mixed model combined with random regression models fitted as a linear polynomial for direct- (a) and maternal additive - (m), as well as permanent environmental (pe) effects. The effect of age at recording was fitted as a cubic spline. Three equally spaced knots per observation were fitted and the same knots were fitted for both the genetic and permanent environmental effects. The fitting of splines and its advantages in random regression analyses are adequately described by Meyer (2005). The model in matrix notation is:
Y = Xβ +Z1a + Z2m +Z3pe + e
where:
• y was a vector of phenotypic observations for weight,
• X was an incidence matrix relating records to the fixed effects (β);
• Z1, Z2 and Z3 were incidence matrices relating records to the direct additive - (Z1), maternal additive -(Z2) and animal permanent environmental effects (Z3)
• a, m, pe and e were vectors of direct additive-, maternal additive-, animal permanent environmentaland residual effects that includes the temporary environmental effects for each observation, respectively.
The model is based on the following assumptions:

where Ka, Km and Kpe are (co)variance matrices between random regression coefficients for direct additive, maternal additive and animal permanent environmental effects, respectively. A is the relationship matrix, I is an identity matrix, NA is the number of animals for which records are available, x is the Kronecker product between matrices and R is a block diagonal matrix containing residual variances. The data were analysed using ASREML (Gilmore et al., 2009).
Results and Discussion
Estimates of direct- and maternal heritability, as well as the ratio due to permanent environmental effects, are depicted in Figure 1.
The direct heritability estimates for the traits were 0.16, 0.13, 0.13, 0.14, 0.16, 0.20, 0.25 for the five traits (BW, WW, YW, FW and 3 MW estimates) respectively. Although the maximum heritability estimate of 0.25 corresponds to results obtained in a recent study on Brangus cattle where multivariate combined with repeatability models were used (Neser et al., 2012), the rest of the values were substantially lower. This is in contrast to results obtained by Meyer (2005) who found, in a study on Angus cattle that heritability estimates from random regression models were higher than results obtained from univariate analysis using the same data. in a study on Nellore cattle, also using random regression models, Albuquerque & Meyer (2001) also reported a slightly lower direct heritability estimates after birth, after which heritability estimates were higher. These estimates are comparable to the current study, especially during the middle stages of recording. Boligon et al. (2010) reported direct heritability estimates that varies from 0.34 (birth) to 0.42 at 550 day of age in Nellore cattle. in another study Arongo et al. (2004) reported direct heritability estimates that varied from 0.38 at 3 years of age to 0.78 at 7 years of age.
The maternal heritability estimates remain low at about 0.05. However, the results for BW correspond to results obtained in a multivariate analysis on Brangus cattle (Neser et al., 2012). The results for WW (0.05 vs. 0.11) were, however, lower. Meyer (2005) also found that the maternal heritability estimates in Angus data using RRM were lower than those found in the literature using other methods of analysis. In a study on Nellore cattle using RRM, Boligon et al. (2010) found that the maternal heritability estimate declined from birth (0.15) to 120 days of age (0.06) and remained constant thereafter.
Conclusion
Sequential culling is the biggest shortcoming in the analysis of growth traits in beef cattle when using RRM, leaving some animals with only one or two records. If this problem could be overcome, accurate modelling of the growth curve of individual animals could ease selection decisions substantially, especially to prevent those cases where selection for heavier weights at early ages could lead to an increase in adult weight.
References
Albuquerque, L.G. & Meyer, K., 2001. Estimates of covariance functions for growth from birth to 630 d of age in Nellore cattle. J. Anim. Sci. 79, 2776-2789. [ Links ]
Arongo, J.A., Cundiff, L.V. & Van Vleck, L.D., 2004. Covariance functions and random regression models for cow weight in beef cattle. J. Anim. Sci. 82, 52-67. [ Links ]
Bohmanova, J. Misztal, I. & Bertrand, J.K., 2005. Studies on multiple trait and random regression models for genetic evaluation of beef cattle for growth. J. Anim. Sci. 83, 62-67. [ Links ]
Boligon, A.A., Mercandante, M.E.Z., Forni, S., Lobo & Albuquerque, L.G., 2010. Covariance functions for body weight from birth to maturity in Nellore cows. J. Anim. Sci. 88, 849-859. [ Links ]
Gilmour, A.R., Cullis, B.R., Welham, S.J. & Thompson, R., 1999. ASREML Reference Manual. NSW Agriculture Biometric Bulletin No.3 NSW Agriculture, Orange Agriculture Institute, Forest Road, Orange 2800 NSW, Australia. [ Links ]
Legarra, A., Misztal, I. & Bertrand, J.K., 2004. Constructing covariance functions for random regression models for growth in Gelbvieh cattle. J. Anim. Sci. 82, 1564-1571. [ Links ]
Meyer, K., 2004. Scope for a random regression model in genetic evaluation of beef cattle for growth. Livest. Prod. Sci. 86, 69-83. [ Links ]
Meyer, K., 2005. Random regression analyses using B-splines to model growth of Australian Angus cattle. Gent Sel. Evol. 37, 473-500. [ Links ]
Misztal, I., Strabel, T., Jamrozik, J., Mantysaari, E.A. & Meuwissen, T.H.E., 2000. Strategies for estimating the parameters needed for different test-day models. J. Dairy Sci. 83, 1125-1134. [ Links ]
Neser, F.W.C., Van Wyk, J.B., Fair, M.D., Lubout, P.C. & Crook, B.J., 2012. Estimation of genetic parameters for growth traits in Brangus cattle. S. Afr. J. Anim. Sci. 42, 469-473. [ Links ]
Nobre, P.R.C., Misztal, I., Tsuruta, S., Bertrand, J.K. Silva, L.O.C. & Lopes, P.S., 2003. Analyses of growth curves of Nellore cattle by multiple-trait and random regression models. J. Anim. Sci. 81, 918-926. [ Links ]
Schaeffer, L.R. & Jamrozik, J., 2008. Random regression models: a longitudinal perspective. J. Anim. Breed. Genet. 125, 145-146. [ Links ]
Copyright resides with the authors in terms of the Creative Commons Attribution 2.5 South African Licence. See: http://creativecommons.org/licenses/by/2.5/za/ Condition of use: The user may copy, distribute, transmit and adapt the work, but must recognise the authors and the South African Journal of Animal Science
# Corresponding author: neserfw@ufs.ac.za














