Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.42 n.3 Pretoria Jan. 2012
Prediction of 305-day milk yield in Brown Swiss cattle using artificial neural networks
O. Gorgulu#
Ahi Evran University, Agriculture Faculty, Department of Animal Science, Biometry and Genetics Unit, 40000, Kirs.ehir /Turkey
ABSTRACT
Artificial neural networks (ANNs) have been shown to be a powerful tool for system modelling in a wide range of applications. In this paper, we focus on the capability of ANNs to predict 305-d milk yield in early lactation of Brown Swiss cattle, based on a few test-day records, and some environmental factors such as age, number of lactation and season of calving. The ANNs that were developed were compared with multiple linear regressions (MLR). The various ANNs were modelled and the best performing number of hidden layers, neurons and training algorithms retained. The best ANN model had input, hidden and output layers of tansig transfer function. The layers had 4, 8, and 1 neurons, respectively. It was determined that the mean predicted values calculated by the ANNs were closer to the real mean values without showing any statistical difference. On the other hand, the predicted mean values calculated by MLR and the real mean values were significantly different from each other. The best prediction in ANN method was seen in 1st, 2nd, 3rd, and 4th test-day records when these were recorded to the system as X1-X8 in the ANN system. In this study, the prediction of 305-d milk yield by ANN gave better results that those of MLR, suggesting that ANN can be used as an alternative prediction tool.
Keywords: Prediction, milk yield, ANN, back propagation, test day records
Introduction
Dairy cattle breeding programmes are based primarily on milk yield and milk composition. Most cows are machine milked twice a day. The official milk records for the herd book are obtained by monthly testing during lactation. Farmers' incomes are derived from milk production and its composition. The accurate measurement or prediction of milk yield is essential to their economy (Fernandez et al., 2007).
Milk yield records that are shorter than the standard lactation should also be used to reduce the bias in estimating breeding values of sires owing to differences in the culling rates among the progeny groups. Early estimates of the sire's breeding value by extending lactations in progress can help to reduce the generation interval, as well as increase the intensity of selection, and thus create greater genetic progress. This early information can allow the farmer to decide whether cows should be kept for breeding. Furthermore, it helps in allocating resources such as feed, both for an individual cow and for a herd (Khan et al., 2005). Lacroix et al. (1995) reported that artificial neural networks (ANNs) allowed for an earlier and a more accurate prediction of milk production in cows. Such improvement is particularly important early in lactation, when a 305-d milk yield can be difficult to predict, and where such a prediction can have serious implications for the choice of future bull-dams. Early detection of low-producing animals is also important for timely culling decisions, with their associated economic benefits (Kominakis et al., 2002). In dairy production, prediction of milk yield is important, in that much of the selection of genetically superior bulls is based on their ability to produce high-yielding daughters. Therefore, the sooner these bulls can be identified, the sooner the collection of semen can commence and insemination of cows can proceed (Sharma et al, 2007).
Artificial neural networks are based on the neural structure of the human brain, which processes information by means of interaction among many neurons. In the past few years there has been a constant increase in interest in neural network modelling in various fields of material science (Taskin et al., 2008). These networks consist of many simple units working in parallel with no central control, and learning takes place by modifying the weights between connections. The basic components of an ANN are neurons, weights and learning rules (Stich et al., 2000). Neurons are organized in layers that process the input information and pass it to the following layer. The processing ability of the network is stored in the inter unit connection strengths (or weights) that are obtained through a process of adaptation to a set of training patterns (Fernandez et al., 2007). Methods based on ANNs seem particularly appropriate in a number of applications, owing to their ability to predict results by learning from the historical data sets of the problem without knowing the interactions among parameters, even if these are highly nonlinear. This ability of ANNs to predict relationships between input variables and their corresponding outputs in a complex biological system has resulted in some inspiring successes (Sharma et al, 2007).
With ANNs, there is no need to begin with an a priori model; nor is there a need to identify all the required variables beforehand. Artificial neural networks have also facilitated the combination of input types (e.g. binary and continuous) and they are potentially advantageous in modelling biological processes that are often characterized as highly non-linear (Lacroix et al, 1995).
In practice, ANNs have been successfully applied in many disciplines, such as engineering, and economic predictions, and in medical diagnoses. There has been relatively little research into the application of ANNs in the field of animal breeding. This is quite paradoxical, as data analyses are usually carried out in this field, and ANNs have shown to be more powerful than classical statistical methods to carry out these kinds of analyses (Fernandez et al, 2006). The reported research has focused on disease detection and dairy cattle breeding, which is concerned with predicting individual milk, fat and protein production. Yang et al. (1999; 2000) applied ANNs to analyses related to predicting clinical mastitis in cattle and found that the technology was able to determine major factors related to the presence or absence of mastitis and to detect influential variables in predicting the incidence of clinical mastitis in dairy cows. Lacroix et al. (1995) and Salehi et al. (1998b) used the networks in milk yield predictions, and demonstrated that adequate preprocessing, a well-designed network model, and a proper set of variables may considerably influence the accuracy of milk production predictions. Salehi et al. (1998a; b) found a neural network model based on back-propagation learning useful in predicting 305-d milk yield, fat and protein. Milk production estimates were successfully obtained in a study by using feed forward ANNs by Sanzogni & Kerr (2001). Artificial neural networks have been applied to predict milk yield in dairy sheep (Salehi et al, 1988). Kominakis et al. (2002) tested the usefulness of ANNs in predicting lactation, as well as daily test milk yield(s) in Chios dairy sheep based on a few (2 - 4) test-day records in the beginning of a lactation period. Grzesiak et al. (2003) compared the neural network and multiple regression predictions for 305-d lactation yield using partial lactation records. Sharma et al. (2007) used an ANN model to predict the first lactation 305-day milk yield using partial lactation records pertaining to Karan Fries crossbred dairy cattle. Hosseinia et al. (2007) estimated second parity milk yield and fat percentage of dairy cows based on first parity information using the neural network system. Njubi et al. (2010) applied ANNs to predict first lactation 305-d milk yield using test-day records in Kenyan Holstein Friesian dairy cows. These studies have shown that total lactation yield and short-term milk yield are positively correlated (Rayalu et al, 1984; Shrivastava et al, 1988; Brutta et al, 1989; Jain et al, 1991; Jadhav et al, 1998).
The aim of this study was to test the usefulness of ANNs in predicting 305-d milk yield, as well as test-day milk yield(s) in Brown Swiss cattle, based on a few (2 - 4) test-day records and some environmental factors (age, number of lactation and season of calving) at the beginning of a lactation period.
Material and Methods
This study was conducted using data on Brown Swiss cattle from Malya State Farm, Kir§ehir, Turkey. Data on 2 640 Brown Swiss cattle were collected each month from July 2002 to January 2009. Usually milk was collected twice daily. The daily milk yield was estimated as the sum of two yields. Season of calving was defined as winter (January to March), spring (April to June), summer (July to September) and autumn (October to December). Statistical analyses were carried out using SPSS 15.0 package. The ANN was designed using MATLAB 7.0.
The design of ANN architecture and methods of training, testing, evaluating and implementing the network is very important. It consists of the choice of ANN algorithm, the structure (number of layers and number of neurons in the layers), the input and output functions, and the learning parameters. This research focuses on the back propagation algorithm learning method. A back propagation algorithm seeks to minimize the error term between the output of the neural net and the actual desired output value. The error term is calculated by comparing the net output with the desired output and is then fedback through the network, causing the synaptic weights to be changed in an effort to minimize error. The process is repeated until the error reaches a minimum value.
Each cow was described with a group of eight variables (X):
X1, average 305-d milk yield
X2, age
X3, number of lactation
X4, season of calving
X5-X8, average test-day milk yield in the 1st, 2nd, 3rd, and 4th month of sampling, respectively.
The data set was divided randomly into three subgroups: the training set (70%), the verification set (15%), and the test set (15%). The verification data set controls the flexibility of the model, preventing model overfitting. It provides a criterion to stop the learning before the model learns the training data, since an excessive adjustment of the model in the training data could lead to poor results when new data are presented to the model. This procedure for model selection is known in the neural network (NN) literature as the early stopping procedure (Fernandez et al., 2007). After fitting the model in the training data, the test data set was used to control whether the processed data set gave a true prediction.
In order to construct the network, the neural network newff function (Equation 1) was used. The constructed network was a back propagation of ANN with three layers of input, hidden and output. The layers had 4, 8 and 1 neurons, respectively. There were seven input (X2, X3, X4, X5, X6, X7, and X8) variables and one output (X1) variable. Figure 1 shows the architecture of the network for the prediction of 305-d milk yield. The tansig transfer function was applied for input, hidden and output layers. The net was trained in 1e+5 cycles of element processing which included epoch = 5000, and goal = 1e-10, where epoch means a single pass through the sequence of all input vectors; and goal means performance is minimized to the goal parameter. In order to study the goodness of the ANN models, some performance indexes took the root mean square error (RMSE) into account.

The MATLAB command newff generates a multilayer layer perceptron (MLP) neural network, which is called net (Beale, 2004; Koivo, 2008).
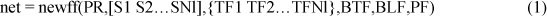
where:
PR - Rx2 matrix of min and max values for R input elements
Si - Size of ith layer
TFi - transfer function of ith layer, default = 'tansig'
BTF - Backprop network training function, default = 'trainlm'
BLF - Backprop weight/bias learning function, default = 'learngdm'
PF - Performance function, default = 'mse'.
Pre-processing data (e.g. standardization and normalization) may lead to an improvement in the learning process of ANN, which helps neural networks to predict better (Hosseinia et al, 2007). We trained NN with normalized data.This normalization keeps the NN from giving primacy to some inputs for their range instead of their importance in solving the problem. Data were normalized at the range -1 to +1 using the premnmx function. After the prediction, in order to see the output, we converted it back to the original scale, using the postmnmx function.
The same data were used for the multiple linear regression (MLR) model. The stepwise regression method was used to obtain the best prediction of the regression model or equation. After the MLR procedure, the least R-squares were obtained. The highest R-squares value was obtained for Equation 2. In this equation, R-squares were calculated as 0.47. Partial regression coefficients were not significant (P >0.05). The MLR was as follows:
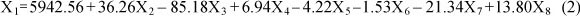
Criteria of goodness of prediction of the ANNs and MLR were Pearson correlation (r) between observed and predicted yields, coefficient of determination (R-squares), standard deviation (σ), the average difference (δ) between observed yields (OY) and predicted yields (PY),

Ratio (ρ) between the standard deviation of differences between observed and predicted yields, and the observed mean value (equation 4) (Kominakis et al., 2002).

Root Mean Square Error (RMSE),

Where, for the ith record, y was the predicted value by ANNs or MLR, yi was the actual value, n was the total number of records (Salehi et al, 1998; Grzesiak et al, 2003).
Ratio of mean (RoM) described by Friedrich et al. (2008) was calculated as:

The difference between the predicted 305-d milk yield values and the real respective values was tested using ANOVA. The significant means were compared using Duncan multiple comparison tests.
Results and Discussion
The results of the statistics of the observed (OY) and predicted (PY) 305-d milk yield for the ANNs and MLR are presented in Table 1. Predicted 305-d mean milk production was very close to that observed for the ANN. However, 305-d milk yield prediction by MLR was lower than the observed 305-d milk yield by 629.4 kg (P <0.01). Grzesiak et al. (2003) used ANN and MLR in their study, showing there was no significant difference between observed values and predicted values (P >0.05) suggesting that ANN was appropriate for modelling 305-d milk yield. The average 305-d milk yield predicted by the ANN was lower than the average observed yield of the 49 reference cows by 13.12 kg (Table 1). The average ANN prognosis did not differ (P >0.85) from the actual average. For the MLR, the average difference was lower by -91.3 kg, but the average yield generated by MLR did not differ (P >0.24) from the actual average yield of the analysed cows (Sanzogni & Kerr, 2001).
Table 1 shows that the highest r value was 0.95 and R2 was 0.90 for the first four test-days when these records were inserted into the net (X1-X8) via ANN module. For 305-d milk yield, the minimum r value was 0.62 and R2, which was 0.38 for X1 -X5 ANN module, while the minimum r value was 0.69 and R2, which was 0.47 for X1-X5 MLR module. Olori et al. (1999) pointed out that R >0.70 implies a very good fit for a model, while R2 <0.40 model should not be used for prediction.
With respect to RMSE values, the minimum value was observed with X1 -X8 ANN. The maximum RMSE value belonged to MLR. Njubi et al. (2010) found similar results to our studies. They obtained R2 = 0.839 and RMSE = 423.3 for ANN module and R2 = 0.676 and RMSE=575.0 for MLR by using the first four testing days.
They concluded that ANN and MLR can both be used for prediction. Sanzogni & Kerr (2001) compared qualitative properties of MLR and two models of ANN. The MLR model, depending on the region, was characterized by an R2 coefficient ranging from 0.78 to 0.86. A classic ANN showed lower R2 coefficients (0.74 - 0.82). The RMS errors for ANN and MLR were similar. In modelling, a high R2 cannot always be a good criterion for prediction. Scatter plots of residuals can be investigated (Alpar, 1997). Figure 2 shows the difference between the observed milk yields and predicted milk yields. According to this graph, the better predictions were obtained for (X1-X7) and (X1-X8) by ANN.
With respect to δ value, the most accurate prediction occurred for X1-X7 ANN. X1-X8 ANN prediction was closer to this prediction. The worst prediction, with regard to δ value, was for MLR. For the ρ value, the best prediction was for X1-X8 ANN. When the ρ value was the highest for X1-X5 ANN module, the MLR prediction on the ρ value was close to that of the ANN module.
With respect to RoM values, ANN modules were similar to each other. The best prediction using 1st, 2nd, 3rd and 4th test records occurred for X1-X8 ANN. This ANN was very close to the ANN used in the 1st, 2nd and 3rd test-days when these were inserted into the system as a variable in X1-X7 ANN. The weakest prediction was seen in the first day of the record inputting X1-X5 ANN. In general, the MLR predictions were weaker than the ANN predictions. Similarly, Sanzogni & Kerr (2001) reported that feed forward ANNs gave better estimates of total milk production than multiple linear regression models, especially when prediction estimates were considered on a regional basis in Australia. Generally, the results encourage further studies on ANNs as an alternative to other biometric methods, with possible use of ANNs for practical on-farm analyses (Grzesiak et al., 2003).
Comparison of results from this study and those from similar studies is important (Schaeffer et al., 2000; Jensen, 2001; Ferreira et al., 2002; Mostert et al., 2006). The use of the test-day model appears to be a better alternative to the 305-d lactation model because early selection based on test-days could reduce generation intervals (Swalve, 1998; 2000; Jensen, 2001) and therefore improve the accuracy of evaluation at farm level. Culling unproductive animals would improve overall farm profitability (Njuibi et al, 2010).
Conclusion
In this paper, (X1 -X8) ANN has been proposed to predict the 305-day milk yield in dairy cattle. The ANN module provided a better prediction for the 305-d milk yield especially when 1st, 2nd, 3rd and 4th test-day records were included as variables into the network. This prediction method was based on the first 4-d milk yield and predicted the real 305-d milk yield, suggesting that this will give animal keepers economic improvements.
References
Alpar, R., 1997. Introduction to Applied Multivariate Statistical Methods I. Nobel Publ., Ankara. (in Turkish). [ Links ]
Beale, M.H., Hagan, M.T. & Demuth, H.B., 2004. Neural Network Toolbox User's Guide (Release 14). The Matwork Inc., Natick MA. [ Links ]
Brutta, S.T. & Pandey, R.S., 1989. Comparative study of ratio and regression methods for predicting lactation yields from part lactation records in dairy cattle. J. Res. Birsa. Agr. U. 1, 117-119. [ Links ]
Fernandez, C., Soria, E., Martin, J.D. & Serrano, A.J., 2006. Neural networks for animal science applications: Two case studies. Expert. Syst. Appl. 31, 444-450. [ Links ]
Fernandez, C., Soria, E., Sanchez-Seiquer, P., Gomez-Chova, L., Magdalena, R., Martin-Guerrero, J.D., Navarro, M.J. & Serrano, A.J., 2007. Weekly milk prediction on dairy goats using neural networks. Neural Comput. Applic. 16, 373-381. [ Links ]
Ferreira, W.J., Teixeira, N.M., Torres, R.A. & Silva, M.V.G.B., 2002. Utilizao da produo de leite no dia do controlena avaliao genetica em gado de leite - Uma reviso. Arch. Latinoam. de Prod. Anim. 10, 46-53. (in Portuguese) [ Links ]
Friedrich, J.O., Adhikari, N.K.J. & Beyene, J., 2008. The ratio of means as an alternative to mean differences for analyzing continuous outcome variables in meta analysis: A simulation study. BMC. Med. Res. Meth. 8, 1-15. [ Links ]
Grzesiak, W., Lacroix, R., Wójcik, J. & Blaszczyk, P., 2003. A comparison of neural network and multiple regression predictions for 305-day lactation yield using partial lactation records. Can. J. Anim. Sci. 83, 307-310. [ Links ]
Hosseinia, P., Edrisi, M., Edriss, M.A. & Nilforooshan, M.A., 2007. Prediction of second parity milk yield and fat percentage of dairy cows based on first parity information using neural networks system. J. Appl. Sci. 7, 3274-3279. [ Links ]
Jadhav, R.G., Vaidya, M.S. & Toro, V.A., 1998. Prediction of first lactation milk yield based on part lactation performance in crossbred cows. J. Bombay Vet. Coll. 6, 57-60. [ Links ]
Jain, A.K., Parmar, O.S. & Gill, G.S., 1991. Efficiency of part lactation milk yield records in progeny testing of Holstein-Friesian bulls. Indian J. Dairy Sci. 44, 479-484. [ Links ]
Jensen, J., 2001. Genetic evaluation of dairy cattle using test day models. J. Dairy Sci. 84, 2803-2812. [ Links ]
Khan, M.S., Hyder, A.U., Bajwa, I.R., Rehman, M.S. & Hassan, F., 2005. Prediction of lactation yield from last-record day and average daily yield in nili-ravi buffaloes. Pakistan Vet. J. 25, 175-178. [ Links ]
Koivo, H.N., 2008. Neural networks: Basics using matlab neural network toolbox. https://noppa.tkk.fi/noppa/kurssi/as-74.3115/luennot/AS-74_3115_neural_networks_-_basics.pdf. (21.10.2010). [ Links ]
Kominakis, A.P., Abas, Z., Maltaris, I. & Rogdakis, E., 2002. A preliminary study of the application of artificial neural networks to prediction of milk yield in dairy sheep. Comput. Electron. Agric. 35, 35-48. [ Links ]
Lacroix, R., Wade, K.M., Kok, R. & Hayes, J.F., 1995. Prediction of cow performance with a connectionist model. Trans. ASAE. 38, 1573-1579. [ Links ]
Mostert, B.E., Theron, H.E., Kanfer, F.H.J. & Van Marle-Köster, E., 2006. Comparison of breeding values and genetic trends for production traits estimated by a lactation model and a fixed regression test-day model. S. Afr. J. Anim. Sci. 36, 71-78. [ Links ]
Njubi, D.M., Wakhungu, J.W. & Badamana, M.S., 2010. Use of test-day records to predict first lactation 305-day milk yield using artificial neural network in Kenyan Holstein-Friesian dairy cows. Trop. Anim. Health Prod. 42, 639-644. [ Links ]
Rayalu, P.K., Subbarayudu, D., Naidu, K.N., Reddy, C.E. & Venkatramaiah, A., 1984. Genetic and non- genetic factors affecting part lactation yields in crossbred cattle. Indian Vet. J. 61, 304-310. [ Links ]
Salehi, F., Lacroix, R. & Wade, K.M., 1998a. Effects of learning parameters and data presentation on the performance of backpropagation networks for milk yield prediction. Trans. ASAE. 41, 253-259. [ Links ]
Salehi, F., Lacroix, R. & Wade, K.M., 1998b. Improving dairy yield predictions through combined record classifiers and specialized artificial neural networks. Comput. Electron. Agric. 20, 199-213. [ Links ]
Sanzogni, L. & Kerr, D., 2001. Milk production estimates using feed forward artificial neural networks. Comput. Electron. Agric. 32, 21-30. [ Links ]
Schaeffer, L.R., Jamrozik, J., Kistemaker, G.J. & Van Doornmaal, B.J., 2000. Experience with a test-day model. J. Dairy Sci. 83, 1135-1144. [ Links ]
Sharma, A.K., Sharma, R.K. & Kasana, H.S., 2007. Prediction of first lactation 305-day milk yield in Karan Fries dairy cattle using ANN modeling. Appl. Soft Comput. 7, 1112-1120. [ Links ]
Shrivastava, G.K. & Khan, F.H., 1988. Relative efficiency of selection based on part production yield in sahiwal herd. Indian J. Dairy Sci. 41, 181-184. [ Links ]
Stich, T.J., Spoerre, J.K. & Velasco, T., 2000. The application of artificial neural networks to monitoring and control of an induction hardening process. J. Ind. Technol. 16, 1-11. [ Links ]
Swalve, H.H., 1998. Use of test day records for genetic evaluation. In: Proceedings of the Sixth World Congress of Genetics Applied to Livestock Production, Armidale, Australia. CD-ROM. [ Links ]
Swalve, H.H., 2000. Theoretical basis and computational for different test-day genetic evaluation methods. J. Dairy Sci. 83, 1115-1124. [ Links ]
Taskin, M., Dikbas, H. & Caligulu, U., 2008. Artificial neural network (ANN) approach to prediction of diffusion bonding behavior (shear strength) of ni-ti alloys manufactured by powder metallurgy method. Math. Comput. Appl. 13, 183-191. [ Links ]
Yang, X.Z., Lacroix, R. & Wade, K.M., 1999. Neural detection of mastitis from dairy herd improvement records. Trans. ASAE. 42, 1063-1071. [ Links ]
Yang, X.Z., Lacroix, R. & Wade, K.M., 2000. Investigation into the production and conformation traits associated with clinical mastitis using artificial neural networks. Can. J. Anim. Sci. 80, 415-426. [ Links ]
Copyright resides with the authors in terms of the Creative Commons Attribution 2.5 South African Licence. See: http://creativecommons. org/licenses/by/2. 5/za Condition of use: The user may copy, distribute, transmit and adapt the work, but must recognise the authors and the South African Journal of Animal Science.
# Corresponding author: ozkangorgulu@gmail.com














