Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Animal Science
versión On-line ISSN 2221-4062
versión impresa ISSN 0375-1589
S. Afr. j. anim. sci. vol.40 no.2 Pretoria ene. 2010
Association of polymorphism of the alpha 1-antitrypsin gene with milk production traits in Chinese Holstein
Qiu-ling LiI; Zheng-feng ZhangII; Chang-fa WangI; Hai YangI; Hong-mei WangI; Jian-bin LiI; Jin-ming HuangI; Ji-feng ZhongI,*
IDairy Cattle Research Centre, Shandong Academy of Agricultural Science, Jinan, Shandong, P. R. China
IIShandong Computer Science Centre, Jinan Shandong, P. R. China
ABSTRACT
Protein degradation in bovine milk affects the quality of dairy products. Alpha 1-antitrypsin (AAT) can protect vulnerable elastic tissues from degradation by neutrophil elastase. The aim of this study was to assess the association of polymorphisms in bovine AAT gene with milk yield and milk composition in Chinese Holstein. Traits analyzed were fat percentage, protein percentage, 305-day milk yield and somatic cell score (SCS). Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), created restriction site-polymerase chain reaction (CRS-PCR) and allele specific-polymerase chain reaction (AS-PCR) methods were used to genotype five loci in coding regions of the sequence, including position 5504, 5609, 5624, 5747 and 8178 in Chinese Holstein. The five mutations were all silent mutation that caused no alteration in the amino acid sequence. In order to determine the relationship between the polymorphisms of the AAT gene and milk production traits and SCS, the General Linear Model (GLM) procedure from the Statistical Analysis Software was used. SNP5504 affected milk fat percentage, SNP8178 affected milk protein percentage and SNP5609 and SNP5624 affected 305-day milk yield. These results suggest that AAT is a candidate gene that influences milk production traits and it could be implemented in breeding programmes to improve the production performance of Chinese Holstein cattle.
Keywords: Dairy cattle, α1-protease inhibitor, SNPs, milk traits, somatic cell score
Introduction
Milk production traits and somatic cell score (SCS) of dairy cattle are important traits that are thought to follow quantitative inheritance. SCS has a high genetic correlation with mastitis with estimates averaging 0.7 (Pösö & Mäntysaari, 1996; Heringstad et al., 2000). Mastitis, inflammation of the mammary glands of dairy animals in both clinical and sub-clinical form results in significant economic losses due to lower milk yields and the milk's degraded quality. Some progress in breed improvement has been made for selection of milk production traits, but they are expensive and time consuming. Contrary to this, marker assisted selection (MAS) can improve the accuracy of selection and therefore genetic progress can be obtained faster and at a lower cost. It is thus very useful to study the genetic variations of candidate genes and their association with milk production and mastitis-related traits (Liefers et al., 2002; Kuss et al., 2003; Yahyaoui et al., 2003; Taylor et al., 2006; Khatib et al., 2007).
Alpha 1-antitrypsin (AAT), a strong protease inhibitor, also known as α1-protease inhibitor (α1PI), belongs to the superfamily of serpins, or serine proteinase inhibitors that include amongst others C1 esterase, antithrombin and α1-antichymotrypsin. AAT is a glycoprotein which forms a SDS staple complex with elastase. The molecular mass of AAT is about 52 kDa, and carbohydrates account for 15% of its mass (Carrell et al., 1982). The bovine AAT gene consists of five exons, spanning about 9 kb of genomic DNA and encoding a 416-AA protein. It is reported that concentrations of AAT in human milk range from 0.1 to 0.6 g/L during the first week of lactation, while concentrations fall during subsequent weeks (Lindberg, 1979; McGilligan et al., 1987; Davidson & Lönnerdal, 1990).
The major role of AAT is to protect tissue against proteolytic digestion by neutrophil elastase (Travis & Salvesen, 1983). In vitro digesting experiments showed that much of the AAT was still intact, while many other milk proteins were digested. The addition of AAT to human milk resulted in a larger proportion of lactoferrin resisting proteolytic degradation (Chowanadisai & Lönnerdal, 2002). It is synthesized primarily in the liver, but is also expressed in, and secreted by, extra-hepatic tissues, intestinal enterocytes, macrophages and monocytes (Perlmutter et al., 1985; Molmenti et al., 1993). Additionally, it has been reported that the AAT protein is produced by the mammary gland and acts as a protease inhibitor in aiding the survival of other biologically active milk proteins. The detection of the transcripts of cDNA derived from the mammary gland supports the suggestion that protease inhibitors are produced locally in the human breast (Urueña et al., 1998). Possible roles of AAT in the immune response include inhibition of lymphocyte toxicity and chemotaxis (Blank & Brantly, 1994). It has been reported that the human AAT gene is associated with a number of human diseases (Majamaa et al., 2001; Lisowska-Myjak & , 2007). Based on the role played by the AAT gene in humans, the aim of this study was to investigate possible associations between variants of the gene and milk production traits as well as SCS in Chinese Holstein dairy cattle.
Materials and Methods
Two hundred and eighty Chinese Holstein cows from seven farms in China were genotyped and analyzed. All blood samples were collected in sterile tubes from the cervical vein of the cattle and mixed with anticoagulant ACD (0.48% citric acid, 1.32% citrate sodium, 1.47% dextrose) at a ratio of 6 : 1 (blood : ACD), and stored at -20 ºC. Genomic DNA was extracted according to the phenol-chloroform method followed by ethanol precipitation (Sambrook et al., 2002). The content of DNA was estimated spectrophotometrically and the genomic DNA was diluted to 50 ng/μL. Milk samples were taken and milk yields recorded once a month for each cow in the course of routine controlled milking over the whole lactation. The fat and protein content as well as the SCC in fresh milk samples were estimated using MilkoScan (FOSS 6000, Denmark). SCS was calculated as SCS = log2 (SCC/100000) + 3, where SCC is cells/μL (Shook, 1993).
The primers used, are presented in Table 1. Primers 1 and 5 that were used to amplify the 448 bp and 370 bp fragments containing mutations G5504A and C8178T have been described by Khatib et al. (2005). Primers 2, 3 and 4 that were used to amplify the 97 bp, 101 bp and 206 bp fragments containing mutation C5609T, G5624T and G5747C were designed based on the bovine AAT gene (GenBank accession number: NC_007319 .2) (Table 1). PCR was carried out in a reaction volume of 20 μL containing 50 ng genomic DNA, 0.5 μmol/L of each primer, 1 × buffer [including 1.5 mmol/L of MgCl2, 100 mmol/L of Tris-HCl, 500 mmol/L of KCl], 0.2 mmol/L of dNTPs and 0.5 U of Taq DNA polymerase. The cycling programme was 4 min at 95 ºC, 35 cycles at 94 ºC for 30 s, X ºC annealing for 30 s, with a final extension at 72 ºC for 7 min (X ºC is annealing temperature, Table 1).
The base underlined is the mismatch introduced. The symbol (+) means there were PCR products by the primers and the symbol (-) means there were no PCR products by the primers (Table 1). G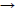 A transition at position 5504, that creates a new Sph I restriction site and C
A transition at position 5504, that creates a new Sph I restriction site and C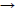 T transition at position 8178, that deletes a Rsa I restriction site, were genotyped with polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). 448 bp and 370 bp PCR products of AAT gene were digested with 5 U Sph I and 5 U Rsa I for 7 h at 37 ºC following the suppliers' directions, respectively. The digested products were detected by electrophoresis in 10% polyacrylamide gel (39 acrylamide: 1 bisacrylamide), respectively.
T transition at position 8178, that deletes a Rsa I restriction site, were genotyped with polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). 448 bp and 370 bp PCR products of AAT gene were digested with 5 U Sph I and 5 U Rsa I for 7 h at 37 ºC following the suppliers' directions, respectively. The digested products were detected by electrophoresis in 10% polyacrylamide gel (39 acrylamide: 1 bisacrylamide), respectively.
The 97 bp and 101 bp fragment containing mutation C5609T and G5624T of the AAT gene were amplified with genomic DNA using Crated Restriction Site-Polymerase Chain Reaction (CRS-PCR) technology. CRS-PCR is a simple and efficient method to identify single nucleotide polymorphisms (SNPs) genotypes. One or more mismatch bases are used in a primer to create a restriction site by combining SNP site after PCR. The CRS-PCR products can be genotyped in the same way as PCR-RFLP. In this study the second base mismatch (A is replaced by C) from 3' end of reverse primer of the second pair of primers with SNP was used to introduce a restriction site after PCR, which formed a restriction site for endonuclease BstFN I. The third base mismatch (T is replaced by C) from 3' end of forward primer of the third pair of primers with SNP, which also creates a new BstFN I restriction site when the bases of positions 5609 and 5624 are wild types (5609-C, 5624-G), respectively, while BstFN I restriction site will be deleted when the bases of positions 5609 and 5624 are mutation types 5609-T, 5624-T, respectively. Four microlitre PCR products of AAT gene fragment were digested with 5 U BstFN I (MBI, Vilnius, Lithuania) for 12 h at 37 ºC following the supplier's directions, respectively. The digested products were detected by electrophoresis in 10% polyacrylamide gel (39 acrylamide : 1 bisacrylamide) and 3% agarose gel, respectively.
The fragment containing a mutation (G5747C) of the AAT gene was amplified with genomic DNA using an allele specific-polymerase chain reaction (AS-PCR). AS-PCR is a single nucleotide mismatch at the 3' end of the annealed reverse primer, which leads the Taq polymerase not to extend the primer. The absenceof the specific PCR product (coupled with a positive internal PCR control) reveals therefore a mutation in the DNA sequence. In each PCR, a forward primer and an AS primer were used in a PCR product from the gene, but failed to produce a product with a mutation at the location covered by the mismatch positions on the AS primer. So, the mutation can be detected by two times PCR only. The AS primers for mutation G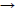 C (at position 5747) detection are shown in Table 1. 206 bp PCR products of the AAT gene fragment were detected by electrophoresis in 1.5% agarose gel.
C (at position 5747) detection are shown in Table 1. 206 bp PCR products of the AAT gene fragment were detected by electrophoresis in 1.5% agarose gel.
Polymorphism information content (PIC), heterozygosity, effective number of alleles and Shannon's information index were calculated using POPGENE Version 1.31 (Yeh et al., 1999).
The General Linear Model (GLM) procedure from SAS (2000) was used to determine the relationship between the polymorphisms of the AAT gene and the investigated traits. Fixed effects of farm, genotype, season of birth, parity and year tested were included as independent variables and the animal's additive genetic effect and permanent environmental effect of individual cows as random effects in the linear model.
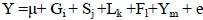 ,
,
where Y is the phenotypic value of milk production traits or SCS; μ is the mean; Gi is the fixed effect of genotype; Sj is the fixed effect of season; Lk is the fixed effect of parity; Fl is the fixed effect of farm; Ym is the fixed effect of test year and eijkl is the random residual effect. The additive and dominance effects of every locus were evaluated separately.
Results
Basic statistics for milk production traits and SCS per farm are shown in Table 2. The five SNPs located in the region of the AAT coding region were genotyped by PCR-RFLP, CRS-PCR and AS-PCR technologies, respectively. Table 3 shows genotypic and allelic frequencies. The heterozygous GA, CT, GT and GC genotypes at positions 5504, 5609, 5624 and 5747 and homozygote CC genotype at position 8178, were the most common genotypes (49.3%, 47.3%, 50.0%, 47.3% and 56.7%, respectively). The alleles G, C, T, C and C were the dominant alleles at positions 5504, 5609, 5624, 5747 and 8178, respectively (0.5069, 0.5757, 0.5115, 0.5069 and 0.6835, respectively).
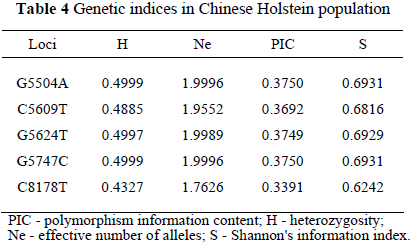
The genetic index evaluated in the Chinese Holstein population was heterozygosities, effective number of alleles, polymorphism information content and Shannon's information index and are presented in Table 4. The genetic index showed that the genetic polymorphisms of these loci were moderate polymorphic loci in Chinese Holstein, according to the criterion of PIC (Vaiman et al., 1994).
Least squares means and standard errors are shown in Table 5 for the effects of the five loci of the AAT gene on milk production traits and SCS. The polymorphisms at G5504A were associated with differences in milk fat percentage (P <0.05). For 305-day milk yield, animals with the homozygous genotype TT at positions 5609 and 5624 had higher 305-day yields than those with the homozygous genotypes, CC and GG (P <0.05). For protein percentage, animals with the homozygous genotype TT had higher milk protein percentages than those with genotypes CC and CT at SNP8178. No other significant associations were observed between the SNPs and milk traits (P >0.05).
Discussion
PCR-RFLP, CRS-PCR and AS-PCR were used to detect the G5504A, C5609T, G5624T, G5747C and C8178T polymorphisms in the AAT gene in order to determine the relationship between these polymorphisms and several milk traits of the Chinese Holstein. The five mutations are all silent mutations that had no alteration in the amino acid sequence, suggesting that the coding region of the AAT gene is relatively conserved. These SNPs were first reported by Khatib et al. (2005), based on direct sequencing of reverse transcription-polymerase chain reaction products in North American Holstein sires. For the bovine AAT gene, there is no other mutation reported. In this study, simple, rapid CRS-PCR and AS-PCR methods were developed to genotype mutations C5609T, G5624T and G5747C. In most loci, the heterozygote was dominant. PIC is a parameter indicative of the degree of informativeness of a marker. The genetic index showed the genetic polymorphisms of these loci to be moderate polymorphic loci in Chinese Holstein (0.25 < PIC < 0.5). Therefore, these loci can be considered useful if they affect the traits of interest.
Results of this study support the conclusion of Khatib et al. (2005) that the AAT gene has association with milk production traits. Previous work, however, did not include the association between these loci and milk traits. Results of the single SNP analysis of this study showed that SNP 5504 affected fat percentage, SNP 8178 affected protein percentage (P <0.05) and SNPs 5609 and 5624 affected 305-day milk yield. Mutations at positions 5504, 5609, 5624 and 8178 have the potential for application in selection programmes. The fat percentage in the AA genotype was higher than that of cows with the GG genotype.
Thus, the allele A (at position 5504) may be associated with an increased fat percentage in the population. The protein percentage of cows with genotype TT was higher than that of cows with genotypes CC and CT. Thus, the allele T (at position 8178) may be associated with an increased protein percentage in this population. The effect of SNPs 5609 and 5624 on 305-day milk yield showed that cows with genotype TT have higher 305-day milk yields than cows with genotypes CC and GG, respectively. Thus, the allele T (at positions 5609 and 5624) may cause an increase in 305-day milk yield of dairy cattle. The genetic effect analysis showed that the AAT gene had a much larger additive than dominance effect.
It is therefore concluded that the AAT gene plays an important role in the process of milk secretion in Chinese Holstein cows. The mutations may change the structure of the AAT protein and thereby milk production traits may be affected; but further verification is needed. The polymorphisms of the AAT gene could be used in breeding programmes as markers to select dairy cattle with increased 305-day milk yield, as well as increased protein and butterfat percentages. However, no significant associations were found between the five mutations in the AAT gene and SCS.
Acknowledgements
The work was supported by 863 High Technology and Development Project of China (2007AA10Z169, 2008AA101010-1), Shandong Project of China (2006YCX028, 2007YCX019, 2007LZ10-06, Y2008D56).
References
Blank, C.A. & Brantly, M., 1994. Clinical features and molecular characteristics of alpha 1- antitrypsin deficiency. Ann. Allergy 72, 105-120. [ Links ]
Carrell, R.W., Jeppsson, J.O., Laurell, C.B., Brennan, S.O., Owen, M.C., Vaughan, L. & Boswell, D.R., 1982. Structure and variation of human alpha-1-antitrypsin. Nature 298, 329-334. [ Links ]
Chowanadisai, W. & Lönnerdal, B., 2002. α1-Antitrypsin and antichymotrypsin in human milk: Origin, concentrations, and stability. Am. J. Clin. Nutr. 76, 828-833. [ Links ]
Davidson, L.A. & Lönnerdal, B., 1990. Fecal alpha-1-antitrypsin in breast-fed infants is derived from human milk and is not indicative of enteric protein loss. Acta Paediatr. Scand. 79, 137-141. [ Links ]
Heringstad, B., Klemetsdal, G. & Ruane, J., 2000. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 64, 95-106. [ Links ]
Khatib, H., Heifetz, E. & Dekkers, J.C.M., 2005. Association of the Protease Inhibitor Gene with production traits in Holstein dairy cattle. J. Dairy Sci. 88, 1208-1213. [ Links ]
Khatib, H ., Zaitoun, I ., Chang, Y.M ., Maltecca, C . & Boettcher, P ., 2007. Evaluation of association between polymorphism within the thyroglobulin gene and milk production traits in dairy cattle. J. Anim. Breed. Genet. 124, 26-28. [ Links ]
Kuss, A.W., Gogol, J. & Geldermann, H., 2003. Association of a polymorphic AP-2 binding site in the 5' flanking region of the bovine β-lactoglobulingene with milk proteins. J. Dairy Sci. 86, 2213-2218. [ Links ]
Liefers, S.C ., te Pas, M.F ., Veerkamp, R.F . & Van der Lende, T ., 2002. Associations between leptin gene polymorphisms and production, live weight, energy balance, feed intake, and fertility in Holstein heifers. J. Dairy Sci. 85, 1633-1638. [ Links ]
Lindberg, T., 1979. Protease inhibitors in human milk. Pediatr. Res. 13, 969-972. [ Links ]
Lisowska-Myjak, B . & Pachecka, J., 2007. Alpha-1-antitrypsin and IgA in serial meconium and faeces of healthy breast-fed newborns. Fetal Diagn. Ther . 22, 116-120. [ Links ]
Majamaa, H ., Aittoniemi, J. & Miettinen, A., 2001. Increased concentration of fecal alpha1-antitrypsin is associated with cow's milk allergy in infants with atopic eczema. Clin. Exp. Allergy 31, 590-592. [ Links ]
McGilligan, K.M., Thomas, D.W. & Eckhert, C.D., 1987. Alpha-1-antitrypsin concentration in human milk. Pediatr. Res. 22, 268-270. [ Links ]
Molmenti, E.P., Perlmutter, D. H. & Rubin, D. C., 1993. Cell-specific expression of a1-antitrypsin in human intestinal epithelium. J. Clin. Invest. 92, 2022-2034. [ Links ]
Perlmutter, D.H., Cole, F.S., Kilbridge, P., Rossing, T.H. & Colten, H.R., 1985. Expression of the a1-proteinase inhibitor gene in human monocytes and macrophages. Proc. Natl. Acad. Sci., USA. 82, 795-799. [ Links ]
Pösö, J. & Mäntysaari, E.A., 1996. Relationships between clinical mastitis, somatic cell score, and production for the first three lactations of Finnish Ayrshire. J. Dairy Sci. 79, 1284-1291. [ Links ]
Sambrook, J. & Russell, D.W., 2002. Translated by Huang Pei Tang. Molecular Cloning A Laboratory Manual. 3nd. Science Press, Beijing, China. [ Links ]
SAS, 2000. Statistical Analysis Software (CD-ROM), Version 8.1, SAS Institute Inc., Cary, N.C., USA. [ Links ]
Shook, G.E., 1993. Genetic improvement of mastitis through selection on somatic cell count, Vet. Clin. Nrth Am. Food Anim. Pract. 9, 563-581. [ Links ]
Taylor, V.J., Beever, D.E., Bryant, M.J. & Wathes, D.C., 2006. Pre-pubertal measurements of the somatotrophic axis as predictors of milk production in Holstein-Friesian dairy cows. Domest. Anim. Endocrinol. 31, 1-18. [ Links ]
Travis, J. & Salvesen, G.S., 1983. Human plasma proteinase inhibitors. Ann. Rev. Biochem. 52, 655-709. [ Links ]
Urueña, C., Telleria, J.J., Blanco-Quiros, A., Arranz, E. & Gomez-Carrasco, J.A., 1998. Alpha-1 antichymotrypsin levels are actively increased in normal colostrum. J. Pediatr. Gastroenterol. Nutr. 26, 376-379. [ Links ]
Vaiman, D., Mercier, D., Moazami-Goudarzi, K., Eggen, A., Ciampolini, R., Lépingle, A., Velmala, R., Kaukinen, J., Varvio, S.L., Martin, P., Levéziel, H. & Guérin, G., 1994. A set of 99 cattle microsatellites characterization synteny mapping and polymorphism. Mamm. Genome 5, 288-297. [ Links ]
Yahyaoui, M.H., Angiolillo, A., Pilla, F., Sanchez, A. & Folch, J.M., 2003. Characterization and genotyping of the Caprine κ-casein variants. J. Dairy Sci. 86, 2715-2720. [ Links ]
Yeh, F.C., Yang, R.C. & Boyle, T., 1999. POPGENE Version 1.31, Microsoft window-based freeware for population genetic analysis. University of Alberta, Canada. [ Links ]
* Corresponding author. E-mail: sdox2@163.com














