Servicios Personalizados
Articulo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en Google
Similares en Google
Compartir
South African Journal of Animal Science
versión On-line ISSN 2221-4062
versión impresa ISSN 0375-1589
S. Afr. j. anim. sci. vol.38 no.4 Pretoria abr. 2008
Genetic diversity and population structure of locally adapted South African chicken lines: implications for conservation
E. van Marle-KösterI; C.A. HeferI; L.H. NelII; M.A.M. GroenenIII
IDepartment of Animal & Wildlife Sciences, University of Pretoria 0002, South Africa
IIDepartment of Microbiology & Plant Pathology, University of Pretoria, Pretoria 0002, South Africa
IIIDepartment of Animal Breeding & Genetics, Wageningen Agricultural University, The Netherlands
ABSTRACT
In this study microsatellite markers were applied to investigate the genetic diversity and population structure of the six local chicken lines kept in the "Fowls for Africa" program, for better clarification of parameters for breed differentiation and genetic conservation of this valuable resource. The lines included the Black Australorp, Potchefstroom Koekoek, New Hampshire, Ovambo, Lebova- Venda and a Naked Neck line. Unbiased estimates for heterozygosity ranged from 50% in the Potchefstroom Koekoek to as high as 65% in the Naked Neck chickens. FIS values varied from as low as 0.16 for the Black Australorp line to as high as 0.35 for the Ovambo chickens. The FST values indicated moderate to high genetic differentiation between the Naked Neck and New Hampshire (0.11); Ovambo and Naked Neck lines (0.12), and Naked Neck and Lebowa- Venda (0.14). A total of 13% of the total genetic variation observed was between the chicken lines and 87% within the lines, supporting moderate genetic differentiation. Population structure was assessed using STRUCTURE where the Black Australorp was genetically best defined. Although six clusters for the different populations could be distinguished, the other lines were not as clearly defined, with individual birds tending to share more than one cluster. Results support a broad classification of these lines and further investigation of unique alleles is recommended for conservation of the lines within the program.
Keywords: Native chicken, microsatellite markers, genetic variation, population structure, South Africa
Introduction
Chickens were introduced to southern Africa during the 1600's by early settlers and traders from Europe and sub-Saharan Africa. Little is known about these early types, but it is believed that their main purpose was the supply of eggs and meat for household consumption (MacDonald & MacDonald, 2000; Ramsey et al., 2000). Various 'more modern' breeds of chickens were introduced from Europe during the era of African colonization, leading to extensive mixing with the established locally adapted chicken lines. The chickens found in rural areas of southern Africa are associated with household food security and are mainly dual-purpose types that have not been subjected to artificial selection in formal breeding programs. These fowl are often only classified based on their phenotype and/or the geographical location and are commonly referred to as "native" (indigenous) birds, distinguishing them from the highly selected modern chicken breeds found in South Africa.
Throughout Africa most research efforts towards the conservation and utilization of indigenous livestock have been focused on cattle and sheep and to a lesser extent on goats (Setshwaelo & Adebambo, 1992, Weigend & Romanov, 2002). Indigenous breeds and strains are particularly vulnerable as selection for improvement in production and uncontrolled mating strategies (inbreeding) may lead to genetic dilution and even loss of genetic variation within these breeds, leading to their eventual extinction (Shresta, 2005; Scherf, et al., 2006). In this regard the locally adapted African chickens were mostly disregarded, and limited data on these breeds have been available in the scientific literature and/or the public domain (Setshwaelo & Adebambo, 1992; Hofmeyr et al., 1998). Although the production potential of the local birds is poor in comparison with the commercial lines, the native breeds have evolved the crucial ability to adapt and survive in often challenging environmental and ecological conditions associated with their geographical origins. Generally, however, exact research data on the genetic potential towards disease resistance and adaptation mechanisms of indigenous livestock and poultry are limited (Anderson, 2003).
In lieu of the above arguments there has been a renewed interest over the past two decades in the conservation and utilization of indigenous livestock, and this has resulted in the establishment of a program for Global Management of Farm Animal Genetic Resources by the Food and Agricultural Organization (FAO). The main objective of this program is to stimulate conservation activities and create an awareness of possible losses of genetic resources on an international scale (Scherf, 1995; Gandini & Oldenbroek, 1999; Shresta, 2005). The information system, Domestic Animal Diversity Information System (DAD-IS), forms one of the main components of the program and provides an inventory of all existing breeds. Several cattle, sheep, goat, pig and horse breeds from 37 European countries are currently listed in DAD-IS (www.fao.org/biodiversity). The biodiversity of 52 chicken breeds from Europe was assessed in a European Union project and a chicken DNA bank and poultry biodiversity database have been established at INRA Jouy-en-Josas (Weigend, 2000).
In South Africa, the Farm Animal Conservation Trust (FACT) was established in 1994 to facilitate and promote conservation of native animal genetic resources. Three South African native chicken lines (Naked Neck, Ovambo and Lebowa-Venda chicken) and one locally developed breed (Potchefstroom Koekoek) have been listed by FACT (Ramsey et al., 2000). The Black Australorp and New Hampshire chickens have been used in South Africa since their importation as dual-purpose breeds during 1925 and 1947, respectively. South Africa has a well developed commercial broiler and layer industry while the locally adapted chicken lines are mainly found in scavenging systems of the rural and peri-urban areas where they are primarily associated with food for household consumption. Here, as in other developing countries, chickens are often associated with the poorer households that cannot afford to keep other livestock such as goats, sheep or cattle (Anderson, 2003). Therefore, despite the existence of the commercial industry, the local chicken lines are found in most villages especially in remote rural areas, where they not only contribute to household food consumption and production, but also play an important role in cultural events.
The need to conserve these local chickens in South Africa was recognized by 1994 and led to the establishment of the "Fowls for Africa" program by the former Animal Improvement Institute, now Agricultural Research Council, Animal Production Institute (API) at Irene (Private Bag X2, Irene 0062). The objectives of this program are to conserve native fowl as a genetic resource and to promote the utilization of these fowl for household food security (Joubert, 1996, Unpublished report). The program was structured to avoid inbreeding and conserve the observed phenotypic differences and genetic variation within the different lines. A phenotypic characterization of the chickens kept in the "Fowls for Africa" program indicated significant differences that contributed to breed definition and the production potential of the various chicken lines (Van Marle-Köster & Casey, 2001). Modern molecular technologies provide the opportunity to study genetic diversity within populations and to identify potentially unique alleles for breed characterization. For conservation of genetic resources, such as those within the "Fowls for Africa" program, it is essential to be able to make informed decisions based on the parameters that describe the genetic diversity and population structure. Among the DNA markers available, microsatellite markers have been shown useful for describing variation and genetic relationships in various farm animal species (Buchanan et al., 1994; Ponsuksili et al., 1996; Zhou & Lamont, 1999; Grzybowski & Prusak, 2004). These markers have generally been found to be highly polymorphic, well suited for diversity studies, and recommended by the FAO Animal Genetic Resources (AGNR). With respect to chickens, a large number of polymorphic microsatellite markers has already been mapped on the chicken genome (Schmid et al., 2000). It was the objective of this study to apply these microsatellite markers for investigating the genetic diversity and population structure among the chickens from the "Fowls for Africa" program, which should lead towards a better clarification of parameters for breed differentiation and genetic conservation of this valuable resource.
Materials and Methods
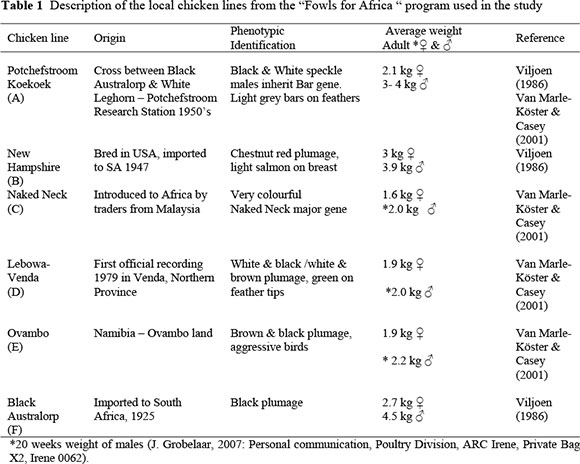
Blood samples for this study were collected from the Potchefstroom Koekoek, New Hampshire, Naked Neck, Lebowa-Venda, Ovambo and Black Australorp, all kept in the "Fowls for Africa" program. These birds were original obtained during 1994 with the establishment of the "Fowls for Africa" program, by J.J. Joubert (Poultry specialist at the former Dairy and Beef Research Institute, Irene), with numbers varying between 100 and 150 for the different groups of chickens. The history of these chicken lines is not well documented. The Lebowa-Venda chickens were first noted by a local veterinarian, Dr Coetzee, who collected these chickens in Thohoyandou area. The other chickens (Naked Neck, New Hampshire and Potchefstroom Koekoek) were collected from rural areas in the Limpopo (former Lebowa and Venda), Free State (former Qwa-Qwa) and Eastern Cape Provinces. The Ovambo birds were collected in Ovambo land in Namibia. No data was available on the specific sampling areas of the original collection for the program (Joubert, 1996, Unpublished report; J. Bester, 2008, Personnel communication, ARC, Private Bag X2, Irene 0062). The Black Australorp formed part of an experimental population at the former Poultry Research Unit at Potchefstroom, and was donated to the ARC, Poultry Division at Irene when the "Fowls for Africa" was established (Joubert, 1996, Unpublished report). A brief description of these chickens included in the study is shown in Table 1. The New Hampshire and Black Australorp are utilized as dual-purpose chickens, and kept in similar systems to the other four lines. Both breeds have been considered as breeds for the upgrading and development of other local lines. These two breeds therefore form part of the "Fowls for Africa" program and were included in the study. According to the origin of these chickens as shown in Table 1, there are three older, established breeds (A, B and F) and three local or native chicken lines (C, D and E).
Between 40 and 60 venous blood samples of each chicken line were collected in 2 mL tubes containing 80 µL EDTA (final concentration 0.5 M). Blood samples were frozen in Eppendorf tubes and kept at -70 °C. DNA was extracted from the blood samples using a Puregene DNA-isolation kit (Gentra Systems, Minneapolis). The concentration of the DNA was determined by spectrophotometry and diluted to a concentration of 10 ng /µL.
Eighteen fluorescently labelled polymorphic microsatellite markers were selected, based on the degree of polymorphism and genome coverage (Crooijmans et al., 1996a; b; Crooijmans et al., 1997). These markers were synthesized and purified at the oligo-nucleotide facilities of the Department of Animal Breeding and Genetics, Wageningen Agricultural University, The Netherlands. Characteristics of these markers are shown in Table 3. PCR reactions were carried out in a volume of 12 µL, containing 10 - 30 ng target DNA, 200 µM dNTP's, 10 mM TrisHCl (pH = 9.0), 1.5 mM MgCl2 0.2 U Taq Polymerase enzyme (Goldstar) and 4 pmoles of each primer (microsatellite marker). Preparation of samples was followed by thermal cycling in a Thermal Controller (Perkin Elmer) using the following program: 5 min at 94 °C followed by 35 cycles consisting of 30 sec at 94 °C, 45 sec at 55 °C, 90 sec at 72 °C and a final extension step of 10 min at 72 °C. GENESCAN-350 TAMRA was used as internal size standard and samples were analyzed on an automated DNA-Sequencer (ABI 377). The Genescan version 2.0 and Genotyper for MacIntosch were used to determine the fragment sizes in base pairs.
Descriptive statistics were calculated to describe genetic diversity (Nei, 1987) with the calculation of the mean number of alleles per locus (MNA), allele frequencies and heterozygosity (expected and observed) and standard deviations using the Microsatellite Toolkit extension to Excel (Park, 2001). GENEPOP version 3.4 was applied for estimation of Fis and Fst statistics (Raymond & Roussete, 1995). The analysis of molecular variance (AMOVA), as implemented by the program, GenAlEx (Peakall & Smouse, 2001), was performed to indicate variation within and between the chicken lines.
For clarification of the population structure of the different lines studied, a Bayesian approach based on the genotypes of the individuals collected and utilizing the software program STRUCTURE (Pritchard et al. , 2000) was used. This involved assigning individuals in the dataset to K (unknown) populations, using a probabilistic approach. K is varied across runs of the program, and individuals have membership assigned to them over all the different clusters (number of clusters = K), where the sum of the probabilities to belong to a population equals one. STRUCTURE was run with 106 iterations, and a burn-in period of 10 000 iterations in order to assure a random starting point for the algorithm. The runs were repeated 20 times for 2 > K < 10, in order to check the consistency of the results. An admixture ancestry model was assumed, which makes provision for the individuals to have a mixed ancestry. This is modelled by assuming that a certain individual (i) has inherited some fraction of its genome from ancestors in population k.
The Factorial Correspondence Analysis (FCA), as presented in the software of GENETIX Version 4 (Belkhir et al., 2004), is a multivariate analysis based on the dissemination of the data according to eigen-vectors as described by She et al. (1987) and Berrebi et al. (2000). This allowed for a graphical representation of the data to discern different groups.
Results
A relatively high degree of polymorphism was observed among the 18 microsatellite markers with the number of alleles per locus ranging from four to 12 (Table 2).
Genetic diversity was expressed as the mean number of alleles (MNA) per locus, percentage polymorphic loci and unbiased heterozygosity (He) for the different lines (Table 3). The MNA alone varied from 3.9 alleles per locus in the Potchefstroom Koekoek to 5.3 in the Naked Neck line. With the exception of the markers LEI 194 and MCW 294 that were monomorphic in the Potchefstroom Koekoek and New Hampshire, all other loci were found to be polymorphic. The genetic diversity (unbiased heterozygosity) He ranged from 0.65 in the Naked Neck population to 0.50 in the Potchefstroom Koekoek, with the average genetic diversity being 0.56.
The FIS values presented in Table 4 indicate the potential inbreeding due to non random mating within the subpopulation while the FST values measure the effects of population subdivision related to potential fixation of alleles relative to the total population (Hartl, 1988). The lowest FIS value (0.16) was observed for the Black Australorp line and the highest value for the Ovambo chickens (0.35). The FST values indicated moderate to high genetic differentiation among the lines; between the Naked Neck and New Hampshire (0.11); between the Ovambo and Naked Neck lines (0.12), and between the Naked Neck and Lebowa-Venda (0.14), according to the guidelines by Hartl (1988).
Variation within and between populations was estimated using AMOVA (Table 5) and the results indicated that a larger proportion (0.87 at a P value = 0.01) of the observed variance occurred within the lines and only 0.13 of the variance contributed to differences between lines.
The population structure of the individual chicken lines was assessed (Pritchard et al., 2000) using all the available data (n = 282). The number of theoretical chicken populations that an individual might belong to (K) varied between two to 10. For the assumed clusters, K = 2, there were two main clusters separating the Black Australorp from the other chicken lines.
In Figure 1, coloured vertical lines (within the rows) represented individual membership to a cluster and the highest proportion of colour therefore indicates that most animals of a specified population (column) were assigned to that cluster. In Figure 1A where K equals 2, only the Black Australorp chickens tend to form a cluster, while there was no clear assignment of the other chicken lines. In the Black Australorp line, where 2 > K < 10, almost all of the individuals showed a high probability of belonging within that specific population. For the LnK estimate, where K = 6 (Figure 1E) the best clustering for the different populations was indicated with some separation among the six lines, but still no clear distinction between the New Hampshire and Lebowa-Venda lines. For the K > 6 < 10 (Figures 1 F-I) there is a trend for separation among the lines but here there were still individuals among the New Hampshire, Naked Neck and Lebowa-Venda that were not clearly defined.
The Factorial Correspondence Analyses (FCA) grouped the lines into six populations as shown in Figure 2, but with a fair amount of overlapping, as is also evident from the Structure analyses, especially between the New Hampshire and the Naked Neck birds.
Discussion
The objective of the study was to investigate the genetic diversity of the different locally adapted chicken lines kept in the "Fowls for Africa" program, for a better understanding of the parameters for breed differentiation and genetic conservation of these lines. There are numerous reasons for conserving genetic variation in indigenous farm animals, including fowl. From the viewpoint of the conservationist, the native fowl should be conserved as a genetic resource against future disasters; commercial chicken stocks are always in danger of severe potential erosion by infectious diseases. Secondly, native fowls could be a source of unique alleles and contribute to the search for genes associated with health and quality traits (Gandini & Oldenbroek, 1999; Mendelsohn, 2003). The different lines from the "Fowls for Africa" program serve both the purposes of conservation of the genetic resource and sustainable utilization. According to K. Fourie (2006, Personnel communication, Private Bag X2, Irene 0062) day-old chickens of all the lines are distributed and marketed to various rural upliftment projects in South Africa. Approximately 36 000 chickens have been hatched and distributed in the NorthWest Province in 2005. The high demand for these chickens due to their potentially higher survival rate under low-input systems is indicative of their importance with regards to both the conservation and the utilization of the resource. Our approach was to use microsatellite markers that were already mapped to the chicken genome and recommended for studies on genetic diversity (Schmid et al., 2000) to investigate genetic variation and population structure of the chickens in the "Fowls for Africa" program. In this way we were able to demonstrate a significant degree of polymorphism and assess the genetic diversity in order to define these chickens as potentially different lines.
In this assessment of the genetic diversity of these chickens the selected microsatellite markers were found to be highly polymorphic with the number of alleles per locus varying from four to 12. This is similar in range to the study on 43 diverse European chicken breeds using 21 microsatellite markers, which included 16 of the markers applied in the present study, where the number of alleles was found to vary between three and 16 (Schmid et al., 2000). In a study on native lines from India and Egypt, the average number of alleles per locus varied between two and 11 (Ponsuksili et al., 1996). In Japanese native breeds an average of 5.6 alleles per locus was reported using microsatellite markers (Takahashi et al., 1998). Studies on commercial chicken lines reported average number of alleles per locus varying between five and three for broilers and layers, respectively (Crooijmans, 1996b).
The heterozygosity (He) values were found to be above 50% for all six of the chicken lines studied, thus indicating a relatively high genetic variability within the groups. The lowest He value of 50% was found in the Potchefstroom Koekoek, followed by the Lebowa Venda and New Hampshire. The high value of 65% heterozygosity in the Naked Neck chickens was expected, partly because classification of chickens into this line was based mainly on the presence of the incompletely dominant Na gene, which is linked to the erythrocyte antigen (CPPP), and which phenotypically results in a chicken with a featherless neck (homozygous) or a little tuft of feathers (heterozygous) (Pitel et al., 2000). No specific breed standards have been specified for the Naked Neck chickens in South Africa. It is therefore possible that chickens carrying the Naked Neck gene from different origins could be classified into this group. The interest in the Naked Neck gene is due to its association with improved heat tolerance (Horst, 1991; Garcês et al., 2001), but has not been studied in the current "Fowls for Africa" program. The Lebowa-Venda is one of the lines considered to be "native" and one could have expected He values similar to that of the Ovambo chicken line. The Lebowa-Venda chickens are associated with a specific geographical area in the former Venda where, according to Joubert (1996, Unpublished report), these chickens were observed by a veterinarian working in the area during the 1970s. They were distinguishable primarily from their very distinct colour pattern. At that time it was reported as the only area in South Africa where these fowls seem to occur. The feathers are speckled white and black or white and brown with green in the sub-layer. The communities in this area probably preferred this pattern and selected them as a group.
In general there is limited data available on genetic profiles of native chickens. In those cases where data have been collected and presented in the literature, the genetic variability varied from as low as 26% to as high as 66%, as was reported in a study on European chicken breeds where microsatellite markers similar to those in the present study were used (Schmid et al., 2000). In a study by Ponsuksili et al. (1996) that included a number of different local breeds, heterozygosity values varied from 33.5% for the Dandarawi and Fayomi (35.1%) from Egypt, 50% for the Nunakan from Indonesia, to as high as 62.9% for the Kadaknath from India. In a more recent study by Muchadeyi et al. (2007) heterozygosity values reported for local chickens were all above 50%; 64 - 66% for Zimbabwe lines, 60.7% for Malawi and 56.1% for Sudan chickens. Genetic variability for commercial broiler and layer lines reported by Groen et al. (1994) was much lower, ranging from 28 to 44%.
The Fis values calculated indicate the potential reduction in heterozygosity due to non-random mating and may serve as an indication of inbreeding within the population (Hartl, 1988). The highest level of potential inbreeding was observed in the Ovambo chickens (0.35) and the lowest in the Black Australorp (0.16). The Fis values for the other local lines were all above 20% and could be due to inbreeding in the original lines sampled, resulting in more related birds than expected. The Black Australorp birds donated to the ARC at Irene, were part of an experimental population from the former Poultry Research Unit at Potchefstroom where mating strategies were in place to prevent inbreeding. This did not occur in the local birds sampled for the conservation program. The sampling of the original lines from different villages could alternatively also indicate potential subpopulations explained by the higher Fis values compared to the Black Australorp coming from one population only.
The subdivision of the lines (FST), as an indication of genetic differentiation among the lines, indicated a moderate to high differentiation among these groups. Muchadeyi et al. (2007), however, reported lower FST values of 0.039 for African chicken lines compared to this study (Table 4). The largest proportion of the total genetic variation (87%) observed, occurred within lines and only 13% between lines. Similar observations were made when microsatellite markers were used to study genetic variation within and between Italian cattle breeds (Ciampolini et al., 1995) and Chinese indigenous pig breeds (Yang et al., 2003). Although genetic differentiation was indicated between the different breeds, variation within breeds was consistently higher.
Due to the fact that local chicken populations are often grouped or described according to their phenotypic characteristics or geographical location, population structure was also assessed for a reflection of the underlying genetic relationships. In this study the Ovambo chickens originate from Ovambo land in Namibia and the Lebowa-Venda from Venda in the Limpopo Province in South Africa and were originally named according to the specific geographical domains. The STRUCTURE (Pritchard et al., 2000) based clustering of the six different lines showed the Black Australorp as genetically best defined, with most of the birds from this line conforming to a distinct group (Figure 1A). For all the other local lines, clusters were not well defined. Although there was a tendency for at least six clusters, the individual birds of these lines tended to assign to more than one cluster. This could be due to genetic similarities between individual birds, and more markers may be required to define these lines. In Figure 2 (FCA analyses) individual birds showed a tendency to conform to six groupings, but still with overlapping between the groups, especially in the New Hampshire and Naked Neck birds. In a study on the phenotypic and production characteristics of these lines, only the Black Australorp and Potchefstroom Koekoek chickens could be distinguished as distinct lines for breed definition (Van Marle-Köster & Casey, 2001). This can be supported by the history of the Koekoek in South Africa, as being bred from a cross between the Black Australorp and White Leghorn during the 1950's, followed by crosses with Plymouth Rock to produce a Black & White speckled bird carrying the sex-linked bar gene (Viljoen, 1986).
The investigation of unique alleles observed in this study could be of importance in the improvement of future strategies towards genetic typing and line identification. A number of private alleles were observed in all lines. Seven such alleles were observed for the Naked Neck, New Hampshire and Ovambo chickens tested, while only one unique allele was observed in the Lebowa-Venda chickens. These alleles can only be confirmed as private alleles with further testing of native chickens in comparison to commercial broiler and layer lines. In a recent review by Muir et al. (2008) on the validation of the 3K chicken SNP array, genetic biodiversity was assessed in commercial broilers and layers. Results indicated a loss of up to 70% in genetic diversity in commercial lines, and confirm the importance of conservation of all potential chicken genetic resources (Muir et al., 2008). Further follow-up studies on the lines from the "Fowls for Africa" program, using more markers, could provide more useful information on the conservation of this genetic resource.
Conclusion
Native fowl types throughout Africa contribute significantly to household food security and are resources with considerable socio-economic value. This study demonstrated that there is moderate to high genetic diversity in the local chicken lines in the "Fowls for Africa" program. Differentiation among the lines is moderate to high and supports a broad classification into separate breeds. Although these lines have been distinguished in terms of phenotype in previous studies, more markers should be added to study the unique alleles and to obtain clarification on defining them as breeds. The "Fowls for Africa" program hosts a valuable genetic resource and it is important that these lines will be conserved as a genetic resource. Genetic information should be taken into consideration in the conservation strategies and in potential upgrading and crossbreeding programs of these lines for rural household food production.
Acknowledgement
The first author wishes to thank the ARC "Fowls for Africa" program for their assistance in obtaining blood samples and interest shown in the project, as well as to N.H. Casey for his valuable support and advice during the initiation and duration of the project in the Department of Animal and Wildlife Sciences, University of Pretoria.
References
Anderson, S., 2003. Analysis: Animal genetic resources and sustainable livelihoods. Ecol. Econ. 45, 331-339. [ Links ]
Belkhir, K., Borsa P., Chikhi L., Raufaste N. & Bonhomme, F., 1996 - 2004. GENETIX 4.05, logiciel sous. Windows TM pour la ganatique des populations. Laboratoire Genome, Populations, Interactions, CNRS UMR 5000, Universit351 de Montpellier II, Montpellier, France. [ Links ]
Berrebi, P., Povz, M., Jesenek, D., Cattaneo-berrebi, G. & Crivelli, A.J., 2000. The genetic diversity of native, stocked and hybrid populations of marble trout in the Soca river, Slovenia. J. Her. 85, 277-287. [ Links ]
Buchanan, F.C., Adams, L.J., Littlejohn, R.P., Maddox, J.F. & Crawford, A.M., 1994. Determination of evolutionary relationships among sheep breeds using microsatellites. Genomics 22, 397-403. [ Links ]
Ciampolini, R., Moazami-Goudarzi, K., Vaiman, D., Dillmann, C., Mazzanti, E., Foulley, J.L., Levesiel, H. Cianci, D., 1995. Individual multilocus genotypes using microsatellite polymorphisms to permit the analyses of the genetic variability within and between Italian beef cattle. J. Anim. Sci. 73, 3259-3268. [ Links ]
Crooijmans, R.P.M.A., 2000. Gene hunting: Molecular analysis of the chicken genome. Doctoral dissertation, Animal Breeding and Genetics group, Department of Animal Sciences, Wageningen University, P.O. Box 338, 6700AH Wageningen, The Netherlands. [ Links ]
Crooijmans, R.P.M.A., Dijkhof, R.J.M., Van der Poel, J.J. & Groenen, M.A.M., 1997. New microsatellite markers in chicken optimized for automated fluorescent genotyping. Anim. Gen. 28, 427-437. [ Links ]
Crooijmans, R.P.M.A., Van Oers, P.A.M., Strijk, J.A., Van der Poel, J.J. & Groenen, M.A.M., 1996a. Preliminary linkage map of the chicken (Gallus domesticus) genome based on microsatellite markers: 77 new markers mapped. Poult. Sci. 75, 746-752. [ Links ]
Crooijmans, R.P.M.A., Groen, A.B.F., Van Kampen, A.J.A., Van der Beek, S., Van der Poel, J.J. & Groenen, M.A.M., 1996b. Microsatellite polymorphism in commercial broiler and layer lines estimated using pooled blood samples. Poult. Sci. 75, 904-909. [ Links ]
Gandini, G.C. & Oldenbroek, J.K., 1999. Choosing the conservation strategy. In: Genebanks and the Conservation of Farm Animal Genetic Resources. Ed. Oldenbroek, J.K., DLO Institute for Animal Science and Health, The Netherlands. [ Links ]
Garcês, A, Casey, N.H. & Horst, P., 2001. Productive performance of naked neck, frizzle and dwarf laying hens under various natural climates and two nutritional treatments. S. Afr. J. Anim. Sci. 31, 174-180. [ Links ]
Goudet, J., 1995. F-stat version 1.2: A computer program to calculate F-statistics. J. Her. 86, 485-486. [ Links ]
Goudet, J., Raymond, M., de Meeüs, T. & Rousset, F., 1996. Testing differentiation in diploid populations. Genetics 144, 1933-1940. [ Links ]
Groen, A.F., Crooijmans, R.P.M.A., Van Kampen, A.J., Van der Beek, S., Van der Poel, J.J. & Groenen, M.A.M., 1994. Microsatellite polymorphism in commercial broiler and layer lines. Proc. 5th World Symp. Gen. Appl. Livest. Prod. 21, 94-97. Guelph, Canada. [ Links ]
Groenen, M.A.M., Crooijmans, R.P.M.A., Veenendal, A., Cheng, H.H., Siwek, M. & Van der Poel, J.J., 1998. A comprehensive microsatellite linkage map of the chicken genome. Genomics 49, 265-274. [ Links ]
Grzybowski, G. & Prusak, B., 2004. Genetic variation in nine European cattle breeds as determined on the basis of microsatellite markers. II Gene migration and genetic distance. Anim. Sci. Papers and Reports 22 (1), 37-44. [ Links ]
Hartl, D.L., 1998. A primer for population genetics, 2nd ed. Sinauer Associates, Inc., Sunderland, MA. ISBN 0878933018. [ Links ]
Hofmeyr, J.H., Bester, J., Fourie, H.J., Scholtz, M.M. & Rege, J.E.O., 1998. Animal genetic resources in Africa. Proc. World Conf. Anim. Prod., Seoul National University, Seoul, Korea. July, 1998. [ Links ]
Horst, P., 1991. Native fowl as a reservoir for genomes and major genes with direct and indirect effects on the adaptability and their potential for tropically orientated breeding plans - A Review, 1991. Animal Research & Dev. Institut für Wissenschaftliche Zusammenarbeit. Federal Republic of Germany. [ Links ]
MacDonald, K.C. & MacDonald, R.H., 2000. The origins and development of domesticated animals in arid West Africa In: The Origins and Development of African Livestock. Eds. Blench, R. & MacDonald, K.C., Ch. 8, 142. Routledge Publishers. Oxon, UK. [ Links ]
Mendelsohn, R., 2003. The challenge of conserving indigenous domesticated animals. Ecol. Econ. 45, 501-510. [ Links ]
Muchadeyi, F.C., Eding, H., Wollny, C.B.A., Groeneveld, S.M., Shamseldin, R., Simianer, H. & Weigend, S., 2007. Absence of population substructuring in Zimbabwe chicken ecotypes inferred using microsatellite analysis. Anim. Genet. 38, 332-339. [ Links ]
Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [ Links ]
Park, S.D.E., 2001. Trypanotolerance in West African cattle and the population genetic effects of Selection. PhD thesis, University of Dublin, Ireland. [ Links ]
Peakall, R. & Smouse, P.E., 2001. GenAlEx V 5: Ggenetic Analysis in Excell. Population genetics for teaching and research. Australian National University, Canberra, Australia. [ Links ]
Pitel, F., Berge, R., Coquerelle, G., Crooijmans, R.P.M.A., Groenen, M.A.M., Vignal, A. & Tixier-Boichard, M., 2000. Mapping the Naked Neck (NA) and Polydactyl (PO) mutants of the chicken with microsatellite molecular markers. Gen. Sel. Evol. 32, 73-86. [ Links ]
Ponsuksili, S., Wimmers, K. & Horst, P., 1996. Genetic variability in chickens using polymorphic microsatellite makers. Thailand J. Agric. Sci. 29, 571-580. [ Links ]
Pritchard, J.K., Stephens, M. & Donnely, P.J., 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945-959. [ Links ]
Ramsey, K., Harris, L. & Kotzé, A., 2000. Landrace Breeds: South Africa's indigenous and locally developed farm animals. Farm Animal Conservation Trust, Pretoria, South Africa www.arc.agric.za [ Links ]
Raymond, M. & Rousset, F., 1995. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Her. 86, 248-249. [ Links ]
Scherf, B.D., 1995. Developing the global inventory for poultry genetic resources. Proc. 3rd Global Conf. Conservation of Domestic Animal Genetic Resources. 1 - 5 August, 1994. Queens University, Canada. Eds Crawford, R.D., Lister, E.E. & Buckley, J.T., Rare Breeds International, Warickshire, UK. [ Links ]
Scherf, B., Rischkowsky, B., Pilling, D. & Hoffmann, I., 2006. The state of the world's animal genetic resources. 8th Wrld Congr. Gen. Appl. to Livest. Prod., 13 - 18 Aug 2006, Belo Horizonte, MG, Brazil. [ Links ]
Schmid, M., Nanda, I., Guttenbach, M., Steinlein, C., Hoehn, M., Schartl, M., Haaf, T., Weigend, S., Fries, R., Buerstedde, J-M., Wimmers, K., Burt, D.W., Smith, J., O'Hara, S., Law, A., Griffin, K., Bumstead, N., Kaufman, J., Thomson, P.A., Burke, T., Groenen, M.A.M., Crooijmans, R.P.M.A., Vignal, A., Fillon, V., Morisson, M., Pitel, F., Tixier-Boichard, M., Ladjali-Mohammedi, K., Hillel, J., Möki-Tanila, A., Cheng, H.H., Delany, M.E., Burnside, J. & Mizuno, S., 2000. First report on chicken genes and chromosomes. Cytogen. Cell Gen. 90 (3 - 4), 169-218. [ Links ]
Schneider, S., Roessli, D. & Excoffier, L., 2000. Arlequin Ver 2.000: A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switserland. [ Links ]
Setshwaelo, L.L. & Adebambo, O.A., 1992. The status and conservation of animal resources in Africa. In: Genetic Conservation of Domestic Livestock. Eds Alderson, L. & Bodó, I., 2, 87-102. CAB International, Farnham, UK. [ Links ]
She, J-X., Autem, M., Kotoulas, G., Pasteur, N. & Bonhomme, F., 1987. Multivariate analysis of genetic exchanges between Solea aegyptiaca and Solea senegalensis (Teleosts, Soleidae). Biol. J. Linnean Soc. 32, 357-371. [ Links ]
Shrestha, J.N.B., 2005. Review article. Conserving domestic animal diversity among composite populations. Small Rumin. Res. 56, 3-20. [ Links ]
Takahashi, H., Nirasawa, Y., Nagamine, Y., Tsudzuki, M. & Yamamoto, Y., 1998. Genetic relationships among Japanese Native breeds of Chicken based on microsatellite DNA polymorphisms. J. Her. 89, 543-546. [ Links ]
Van Marle-Köster, E. & Casey, N.H., 2001. Phenotypic characterization of native chicken lines in South Africa. Anim. Genet. Res. Info. 29, 71-78. FAO, Rome, Italy. [ Links ]
Viljoen, W.C.J., 1986. Hoenderrasse. Pluimvee A2. Boerdery in Suid-Afrika. Department of Agriculture, Pretoria 0002, South Africa. [ Links ]
Weigend, S., 2000. Final report: Development of strategy and application of molecular tools to assess biodiversity in chicken genetic resources (AVIAN DIV). Institute for Animal Science and Animal Behaviour, Federal Agricultural Research Centre, Braunschweig, Germany 31535 Neustadt. [ Links ]
Weigend, S. & Romanov, M.N., 2002. The world watch for domestic animal diversity in the context of conservation and utilization of poultry biodiversity. Wrld Poult. Sci. J. 58, 411-430. [ Links ]
Yang, S., Wang, Z., Liu.B., Zhang, G., Zhao, S., Yu, M., Fan, B., Li, M. & Li, K., 2003. Genetic variation and relationships of eighteen Chinese indigenous pig breeds. Genet. Sel. Evol. 35, 657-671. [ Links ]
Zhou, H. & Lamont, S.J., 1999. Genetic characterization of biodiversity in highly inbred chicken lines by microsatellite markers. Anim. Gen. 30, 256-264. [ Links ]
 Correspondence:
Correspondence:
E-mail: evm.koster@up.ac.za














