Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.35 n.4 Pretoria 2005
SHORT COMMUNICATION
Angoradb: A database for QTL research in Angora goats
C.A. HeferI; F. JoubertII; E. van Marle-KösterI, #
IDepartment of Animal and Wildlife Sciences, University of Pretoria, Pretoria 0002, South Africa
IIBioinformatics and Computational Biology Unit, Department of Biochemistry, University of Pretoria, Pretoria 0002, South Africa
ABSTRACT
Biological data repositories are widely applied to assist researchers in effectively storing and retrieving data. The recent developments in molecular biology intensified the need for well-designed biological databases, which can be queried without increasing the computational time. This study describes the design of a biological databank for the primary data collection phase, which involves collection and storing of genotypic and phenotypic information of a number of South African Angora goat herds, which will be utilised in a QTL study for mohair traits.
Keywords: Angora goats, biological databank, genotypic, phenotypic information
The recent exponential growth in biological data due to developments in molecular genetics and genomics has intensified the need for well-designed biological databases, which can be queried without increasing the computational time required to perform queries. Major drawbacks in the design of biological data repositories are the level of domain specific knowledge needed to understand the data, and the descriptive nature of biological data (Kasprzyk et al., 2004).
Economically important traits are mostly under control of quantitative trait loci (QTL), which need to be mapped and identified in order to provide a knowledge base and to be applied as a tool for genetic improvement of farm animals. The basic principle of QTL mapping lies in the association of the measurable genetic variation with the phenotypic variation, which occurs within a family or pedigree of animals. QTL mapping relies heavily on sound experimental design, efficient data storage and analyses (Bovenhuis et al., 1997; Sonstegard et al., 2001). Genotypic and phenotypic information stored together in an integrated format is an essential step for successful QTL experiments. The detection of QTLs is underway in various commercial animal populations, with QTLs being detected for a number of traits. Traits include for example meat and carcass quality of pigs (Ciobanu et al., 2004; Thomsen et al., 2004, Milan et al., 2002), beef cattle (Li et al., 2004; Cases et al., 2003) and chickens (Ikeobi et al., 2002) and milk, protein and butterfat yield in dairy cattle (Jiang et al., 2005). Goats in general are poorly researched when compared to sheep and cattle. QTL research in Angora goats has mostly been limited to certain growth traits and body measurements (Marrube et al., 2004).
This study describes the design of a biological databank for the primary data collection phase of both genotypic and phenotypic information of animals genotyped in a fine hair flock of Angora goats, which will be utilised in a QTL study for mohair traits.
The Angora fine hair flock at the Jansenville Experimental Station, in the Eastern Cape province of South Africa, formed the reference population for constructing the database for this study. Stud breeders taking part in the official recording scheme for small stock were approached and four breeders will also be contributing to the database.
The open source database management system (DBMS) POSTGRESQL was used as the backbone for construction of the database. CGI scripts were used to establish a communication with the HTML front end. PERL (version 5) scripts handled user restriction, verification and security issues, and session control was performed via CGI cookies (Figure 1). PERL was also used to construct and perform the queries to the DBMS, and retrieve and parse the query results.
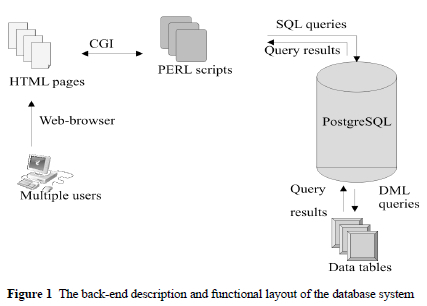
Batch uploads of data, as well as single entry uploads to the database are possible, and the results of queries can be sent via e-mail as a comma delimited flat file in order to import the information into popular word processing software suites. Strict rules exist when uploading data into the database to ensure referential integrity of the data, with templates being made available for batch uploading.
Pedigree information and performance data of traits, namely fleece weight, fibre characteristics (fibre diameter, staple length and proportion of medullated fibres) and body weight at weaning, year old and 18 months of age are recorded by the stud breeders taking part in the Small Stock Improvement Scheme in South Africa. The database, consisting of eight tables, was designed to group the information into three sections, namely phenotypic, genotypic and general information of the animal, user and the breeder (Figure 2). Genotypic information will provide information on the blood samples obtained from the breeder, DNA extraction procedures and microsatellite marker analyses performed for the various flocks.
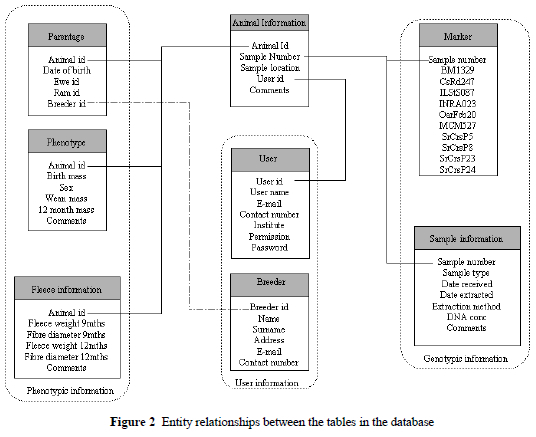
Access to the database is made possible via web-based html pages, which allow for access control. Restricted access is provided to allow breeders involved in the project to view and query data, with additional permissions granted to the researchers involved in submitting and curating the datasets.
The databank forms a primary repository for the collection and storage of data for the QTL study. Genotypic information, together with phenotypic and parentage information can be stored in the databank, providing a framework for the research involved in the QTL detection project. This databank has the potential to become the major repository of fine hair trait information of Angora goats in South Africa, and the detection of actual QTL involved in controlling these traits will attest to the success of this data repository. Future developments of this databank into a full scale QTL data repository and viewer, such as the bovine QTL viewer (http://bovineqtl.tamu.edu/) are a possibility, depending on the success of actual mapping of mohair trait QTL to the Angora genome.
References
Bovenhuis, H., Van Arendonk, J.A.M., Davis, G., Elsen, J.M., Haley, C.S., Hill, W.G., Baret, P.V., Hetzel, D.J.S. & Nicholas, F.W., 1997. Detection and mapping of quantitative trait loci in farm animals. Livest. Prod. Sci. 52:135-144. [ Links ]
Cases, E., Shackleford, S.D., Keele, J.W., Koohmaraie, M., Smith, T.P.L. & Stone, R.T., 2003. Detection of quantitative trait loci for growth and carcass composition in cattle. J. Anim. Sci. 81, 2976-2983. [ Links ]
Ciobanu, D.C., Bastiaansen, J.W.M., Lonergan, S.M., Thomsen, H., Dekkers, J.C.M. & Rothschild, M.F., 2004. New alleles in calpastatin gene are associated with meat quality traits in pigs. J. Anim. Sci. 82, 2829-2839. [ Links ]
Ikeobi, C.O.N., Wooliams, J.A., Morrice, D.R., Law, A., Windsor, D., Burt, D.W. & Hocking, P.M., 2002. Quantitative trail loci affecting fatness in the chicken. Anim. Gen. 34, 428-435. [ Links ]
Jiang, Z., De, S., Garcia, M.D., Griffin, K.B., Wu, X.L., Xiao, Q., Michal, J.J., Sharma, B.S. & Jansen, G.B., 2005. An independent confirmation of a quantitative trait locus for milk yield and composition traits on bovine chromosome 26. J. Anim. Breed. Gen. 122 (4), 281-284. [ Links ]
Kasprzyk, A., Keefe, D., Smedley, D., London, D., Spooner, W., Melsopp, C., Hammond, M., Rocca-Serra, P., Cox, T. & Birney, E., 2004. EnsMart: A Genetic System for fast and flexible access to biological data. Genome Res. 14, 160-169. [ Links ]
Li, C., Basarab, J., Snelling, W.M., Benkel, B., Kneeland, J., Murdoch, B. & Moore, S.S., 2004. Identification and fine mapping of quantitative trait loci for back fat on bovine chromosomes 2,5,6,19,21 & 23 in a commercial line of Bos taurus. J. Anim. Sci. 82, 967-972. [ Links ]
Marrube, G., Cano, E.M., Roldan, D.L., Abadm, M., Allain, D., Vaiman, D., Taddeo, H. & Pilo, M.A., 2004. Preliminary results of a genome scan for QTLs affecting growth in Angora goats. 8th Internat. Conf. on Goats, South Africa, July 2004, 24. [ Links ]
Milan, D., Bidanel, J., Iannoccelli, N., Riquet, J., Amiques, Y., Gruand, J., Le Roy, P., Renard, C. & Chevalet, C., 2002. Detection of quantitative trait loci for carcass composition traits in pigs. Gen. Sel. Evol. 34, 705-728. [ Links ]
Sonstegard, T.S., Van Tassel, C.P. & Aswell, M.S., 2001. Dairy cattle genomics: Tools to accelerate genetic improvement? J. Anim. Sci. 79, (E Suppl.) E307-315. [ Links ]
Thomson, H., Lee, H.K., Rothschild, M.F., Malek, M. & Dekkers, J.C.M., 2004. Characterisation of quantitative trait loci for growth and meat quality in a cross between commercial reeds of swine. J. Anim. Sci. 82, 2213-2228. [ Links ]
# Corresponding author. E-mail: evm.koster@up.ac.za














