Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
South African Journal of Animal Science
On-line version ISSN 2221-4062
Print version ISSN 0375-1589
S. Afr. j. anim. sci. vol.33 n.3 Pretoria 2003
Molecular genetic analysis of the Chinese Erhualian pig breed
G. H. YueI, II, ; G. L. WangI
ICollege of Animal Sciences and Technology, Nanjing Agricultural University, 210095 Nanjing, China
IICurrent address: Temasek Life Sciences Laboratory, 117604 Singapore
ABSTRACT
The Chinese Erhualian is one of the most prolific pig breeds in the world, but it is in danger of being replaced by other exotic pig breeds because of its slow growth rate and high fat content in the body. To obtain some genetic information for conservation, we analysed the Erhualian pigs by using a PCR-RFLP for the calcium-release-channel (CRC) gene, nine polymorphic microsatellites and the complete mtDNA D-loop sequences, and compared these data with those from other pig breeds from Europe and Asian. The PCR-RFLP analysis of the CRC gene showed that the frequency of the C allele associated with stress resistance was 100% in the Erhualian pigs. Neighbour-Joining trees constructed on the basis of mtDNA D-loop sequences and the microsatellite analysis clearly showed that the Erhualian pigs were located in a separate branch. These data suggest that the Erhualian pigs are different from other breeds. Microsatellite analysis showed that the average allele number (5.3/locus) in the Erhualian pig was intermediate as compared with that (4.8-7.0/locus) in the three European pig breeds. The expected heterozygosity was higher in the Erhualian pig (0.78) than that in these European pig breeds (0.59-0.72), whereas the observed heterozygosity was higher in the European breeds (0.51-0.64) than in the Erhualian pig (0.46). In the Erhualian pig, the fixation index (FIS) was as high as 0.41. These data suggest a high level of inbreeding and/or subpopulation in the Erhualian pigs. For conservation of the germplasm in the Erhualian pigs, it is necessary to take measures to reduce inbreeding and/or subpopulation.
Keywords: Erhualian pig, genetic diversity, conservation, DNA markers
Introduction
The Erhualian pig located in the Jiangsu and Zhejiang provinces is one of the 48 native Chinese pig breeds (Zhang, 1986). Erhualian pigs can use a high proportion of forage foods in their diets, and offer valuable traits (e.g. early sexual maturity, large litter size, tasty meat and disease resistance) for pig breeders world-wide (Zhang, 1986). The Erhualian is the most prolific pig breed in China, and among the most prolific breeds in the world (Zhang, 1986, Rothschild & Ruvinsky, 1998). The male pigs reach sexual maturity at the average age of 90 days, with a mean body weight of 15.6 kg, whereas the females reach sexual maturity at the age of 64 days, with an average body weight of 15.0 kg. The litter size of 278 sows averaged 15.93 (Yue, 1994). Currently some Chinese native breeds are in danger of being replaced by exotic breeds with very different feed requirements, reproduction rates and meat qualities mainly because of the slow growth rate and high fat content in body in the Chinese breeds (Chang et al., 1999)). The Erhualian has been listed under the category of protected breeds by the Chinese Ministry of Agriculture (Chang et al., 1999).
The Erhualian is closely related to the Meishan pig breed (Zhang, 1986), which is now well known in Europe and in the USA (Rothschild & Ruvinsky, 1998). Several Meishan pigs were exported to the USA and Europe in 1980s, and had been used in breeding programs (Ducos et al., 1992) and in pig genome mapping projects (Geldermann et al., 1996; Rohrer et al., 1996). A study on QTL mapping showed that the Meishan pig possessed QTL alleles which could reduce fat content in carcass, despite its fat appearance (Yue et al., 2003). Research efforts in Europe and the USA have been aimed at explaining the mechanisms underlying the high fecundity and lower embryonic mortality of the Meishan pigs (Rothschild & Ruvinsky, 1998). A study carried out in the USA showed that the allelic diversity of the imported Meishan pigs was higher than that of some commercial breeds (Paszek et al., 1998). A PCR-RFLP analysis of calcium-release-channel (CRC) gene showed that the frequency of the T allele associated with stress susceptibility was zero in Meishan pigs (Knorr et al., 1994). Studies on genetic diversity have been carried out for a number of breeds (e.g. Paszek et al., 1998; Laval et al., 2000; Kim & Choi, 2002). However, molecular studies on the Erhualian pigs are just beginning (Fan et al., 2002; Jiang et al., 2002). There is little detailed information on allele frequencies at the CRC locus, the genetic diversity and genetic relationship of the Erhualian pigs with other pig breeds.
In this study, we analysed the Chinese Erhualian pigs by using three different DNA marker systems, i.e. a PCR-RFLP for the CRC gene, nine polymorphic microsatellites and the complete mtDNA D-loop sequences, and compared the data from the Erhualian pigs with data from other European and Asian pig breeds. The main purpose is to supply some basic information for conservation and application of the prolific Erhualian pigs.
Materials and Methods
Blood samples of 32 unrelated Erhualian pigs and two unrelated Meishan pigs were collected in Jiangsu (22 individuals: 12 females and 10 males) and Zhejiang (10 individuals: 6 females and 4 males) provinces, China. DNA was isolated from whole blood as described (Miller et al., 1988). DNA samples of 30 to 40 individuals from each of the three European breeds (Landrace, Pietrain and Large White) were obtained from University of Hohenheim, Germany (Chen, 1994; Laval et al., 2000).
A PCR-RFLP for the CRC gene was performed only for the Erhualian pigs according to Knorr et al. (1994), since genotype data at the CRC locus are available in other breeds studied here (Knorr et al., 1994). Briefly, a 655 bp fragment containing the point mutation (C to T) in the relative position +1843 of the CRC gene (Fujii et al., 1991) was amplified by using a primer pair (5' GTTTGCCACAGGTCCTACCA 3', and 5' ATTCACCGGAGTGGAGTCTC 3'). The amplified fragment was then digested by the use of the HinP I (New England Biolab), leading to two fragments: 489 bp and 166 bp for the C/C genotype, and an undigested band of 655 bp for the T/T genotype.
To amplify the complete mtDNA D-loop sequences of pigs, one primer pair (5' CAACCAAAACAAGCATTCCATTCGT 3' and 5' CGCCCACAAT-CTGAATATAGCTTATA 3') was designed according to the known pig D-loop sequences (GenBank No. AF276930 and AF276928). PCR was conducted on a PCR-machine PTC-200 (MJ Research) using 50 ng genomic DNA in a 25 μl reaction volume containing 0.4 μΜ of each primer, 2 00 μΜ of dNTPs, 1x PCR buffer with 1.5 mM MgCl2 (Finnzymes) and 0.5 U of DNA-polymerase (Finnzymes). The PCR conditions were: 94 °C for 2 min, and then 35 cycles at 94 °C for 30 sec, 55 °C for 30 sec and 72 °C for one minute followed by an extension at 72 °C for 5 min. The PCR products of about 1044 bp were cloned into a pGEM-T vector (Promega), then sequenced using M13 or M13 reverse primers using BigDye chemicals (ABI/PE) as described (Yue et al., 2000). Forward and reverse sequences were aligned by using Megalign in DNASTAR software package (DNASTAR Inc.). Amplified PCR-products from four Erhualian (Erhualian01-Erhualian04) and two Meishan (Meshan01 and Meishan02) individuals were sequenced, whereas for the three European pig breeds, one individual from each breed was sequenced. The mtDNA D-loop sequences of these five breeds (Erhualian, Meishan, Large White, Landrace and Pietrain) were deposited in the GenBank (accession number: from AY230818 to AY230827). For a phylogenetic analysis, known sequences of the complete mtDNA D-loop of other pig breeds were also used. These breeds included eight Chinese breeds (Nanjang, Wanahua, Gonghei, Dongchang, Jinghua, Putian, Ningxian and Erhualian), a Korean breed (Cheju), two Japanese wild boars (JP-wild01 and JP-wild02) and two European breeds (one UK Walsh and two European wild boars: Eu-wild01 and Eu-wild02). The D-loop sequence of the fifth Erhualian individual (Erhualian05) was derived from GeneBank. Sequences were aligned by using the ClustalX 1.8 (Thompson et al., 1997). Gaps in the aligned sequences were treated as missing. Sequence divergence was calculated using the software MEGA (Kumar et al., 2001). A Neighbour-Joining (NJ) tree was constructed using the MEGA (Kumar et al., 2001). The confidence of branching order was estimated by the use of the bootstrap analysis using 1000 replicates.
Nine microsatellites of 27 microsatellites approved for studying porcine genetic diversity by the FAO-ISAG advisory committee (Laval et al., 2000) were selected for genotyping all the Erhualian, Large White and Pietrain pigs. These nine markers are S0155 (located on chromosome 1: SSC1), Sw240 (SSC2), S0226 (SSC2), S0002 (SSC3), IGF-I (SSC5), Sw951 (SSC10), S0090 (SSC 12), Sw857 (SSC14) and S0026 (SSC16). One primer of each pair was labelled with a fluorescent dye (either Hex or Fam or Tet). Genotyping of microsatellites was carried out using either an automated A.L.F. DNA sequencer (Pharmacia) or an ABI 377 DNA sequencer as described previously (Yue et al., 1999; Yue et al., 2000). Genotype data of the German Landrace were obtained from an earlier study (Laval et al., 2000) and used for comparison.
The number and frequency of alleles at microsatellite loci, Wright's fixation indices (FIS and Fst) and expected (HE) and observed (HO) heterozygosity were calculated using the GDA software package (Lewis & Zaykin, 2000). An exact test was carried out to examine the Hardy-Weinberg Equilibrium using the GDA (Lewis & Zaykin, 2000).
The (δμ)2 genetic distance (Goldstein et al., 1995) and Nei's standard genetic distance (Nei, 1978) were computed by using the Microsat program (Minch, 1996). The (δ/)2 genetic distance was then adjusted for small population size (Goldstein & Pollock, 1997). The NJ trees were constructed based on the two genetic distance measures by the use of the MEGA software package (Kumar et al., 2001).
Results
The PCR-RFLP analysis of the CRC gene revealed that all 32 Erhualian pigs had the same genotype C/C, indicating the frequency of the C allele associated with stress resistance was 100% in the Erhualian pigs.
The genetic diversity of the Erhualian pigs is summarised in Tables 1 and 2. At nine microsatellite loci, a total of 48 alleles were detected in the Erhualian pigs. This gives an average number of 5.3 alleles per locus. The allele number of the Erhualian was less than that of the Large white, Pietrain, but higher than that of the Landrace. A total of 15 private alleles was detected in the four breeds. The Large White and Pietrain breeds displayed most private alleles (each had seven private alleles), The Erhulian pigs exhibited one (the allele 120 bp at the locus Sw951), whereas the Landrace showed none. In the Erhualian, disequilibrium of the genotypic frequencies (P < 0.05) was found at eight loci except at the locus S0026, whereas in the European pig breeds disequilibrium of genotypes was seen at three to six loci (Table 1).
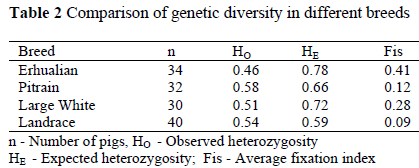
The mean observed heterozygosity (HO) at all nine microsatellite loci ranged from 0.46 for Erhualian to 0.58 for Pietrain. The expected heterozygosity (HE) was higher in the Erhualian than in European breeds. The fixation index (FIS) ranged from 0.12 for Pietrain to 0.41 for the Erhualian (Table 2). The average Fst between breeds was 0.11. All Fst calculated by pairs of breeds were different from zero (P < 0.05).
A phylogenetic analysis of mtDNA D-loop sequences revealed two distinct clusters, one for Asian breeds (A) and the other for European breeds (B) (Figure 1). Within the cluster of the Asian breeds, the Erhualian pigs were located on a separate branch, which was near the branch of the two Meishan and the Nanjing pigs. The average pairwise nucleotide distance between the Asian and European breeds was 1.9%.
Based on microsatellite data, the (δμ)2 and Nei's standard genetic distance measures showed the smallest genetic distance was between Large white and Pietrain, and the longest distance between Erhualian and Landrace (data not shown). The genetic distance between the Erhualian and European breeds was much larger than that between different European breeds. Clearly independent cluster of the Erhualian from the European breeds was seen in both trees (Figure 2).
Discussion
The calcium-release-channel (CRC) gene is an important candidate for pig malignant hyperthermia, which is an inherited skeletal muscle disorder causing major economic losses in the pig industry. A point mutation at the +1843 of the CRC gene (C to T) is the major cause of stress susceptibility (Fujii et al., 1991). DNA tests on the point mutation in several pig breeds clearly demonstrated that the T allele associated with stress susceptibility existed in almost all species tested (Grobet et al.; 1992; Houde et al;, 1993; Knorr et al., 1994), except the Chinese Meishan pig. In this study we did not find the T allele in the Erhualian pigs, suggesting the Erhualian pigs are free of stress susceptibility. Removal of the T allele from pig populations through traditional selection, even with marker assisted selection is time consuming and costly. Therefore, besides the high reproductive performances, high meat quality and disease resistance, the Erhualian would offer another good trait (i.e. stress resistance) for pig breeders world-wide.
The phylogenetic trees constructed on the basis of complete mtDNA D-loop sequences and the microsatellite analysis showed a marked genetic difference between Asian breeds and European breeds. This result is consistent with those of other studies on the evolutionary relationships between Chinese and Western pig breeds (Li & Enfield, 1989; Chang et al., 1999; Giuffra et al., 2000). Within the cluster of Asian pig breeds, Erhualian pigs were located in a separate branch in the tree constructed by the use of mtDNA D-loop sequences, suggesting the Erhualian breed is genetically different from not only European pig breeds, but also other Chinese native breeds.
The allele number of the Erhualian was less than that of the Large White and Pietrain, but higher than that of German Landrace. As compared with the published data (Laval et al., 2000; Li et al., 2000; Lemus-Flores et al., 2001; Kim & Choi, 2002), the average allele number of the Erhualian was higher than most pig breeds studied. Since the average number of alleles per locus is sensitive to the sample size and microsatellites used, expected and observed heterozygosity should be preferred for comparisons. The Chinese Erhualian showed higher expected heterozygosity than some published data on pigs (Laval et al., 2000; Kim & Choi, 2002), suggesting higher gene diversity in the Erhualian pigs, which correspondents to the results on the Meishan pigs (Paszek et al., 1998). The observed heterozygosity was higher in other pig breeds (Kim & Choi, 2002; Laval et al., 2000) than in the Chinese breed. The fixation index FIS, which is sensitive to heterozygote deficit, was as high as 0.41 in the Chinese breed, suggesting a high level of inbreeding and/or subpopulation in the Erhualian breed and/or appearance of null alleles at microsatellite loci. But the possibility of appearance of null alleles at the nine microsatellites used here can almost be ruled out, because the nine microsatellites were selected from more than 400 microsatellites from linkage maps (Archibald et al., 1995; Rohrer et al., 1996). They have also been used in several genetic diversity studies (Laval et al., 2000; Li et al., 2000), and none has detected null alleles. The deviation from the Hardy-Weinberg Equilibrium at most loci in the Erhualian pigs also suggests subpopulation within this breed. For conservation of Erhualian pigs, measures should be taken to reduce inbreeding and/or subpopulation. Microsatellites could be used to analyse the relatedness of individuals and to arrange the mating pairs properly for reducing inbreeding. It is also useful to exchange some breeding boars from different regions. Genotypes at polymorphic microsatellite loci could help to identify which individuals should be exchanged between different locations.
Conclusion
The Erhualian breed is quite different from other pig breeds, and could offer valuable traits, such as high reproductive performance, disease resistance, stress resistance and high meat quality for pig breeders world-wide. Therefore, its germplasm is worth to be conserved. However, high level of inbreeding and/or subpopulation have been detected in this study. Thus for conservation of this valuable germplasm, measures should be taken to reduce inbreeding and subpopulation.
Acknowledgement
The authors would like to thank Z.H. Jiang, Y.Z. Chen and H. Geldermann for supplying DNA samples.
References
Archibald, A.L., Haley, C.S., Brown, J.F., Couperwhite, S., Mcqueen, H.A., Nicholson, D., Coppieters, W., Vandeweghe, A., Stratil, A., Wintero, A.K., Fredholm, M., Larsen, N.J., Nielsen, V.H., Milan, D., Woloszyn, N., Robic, A., Dalens, M., Riquet, J., Gellin, J., Caritez, J.C., Burgaud, G., Ollivier, L., Bidanel, J.P., Vaiman, M., Renard, C., Geldermann, H., Davoli, R., Ruyter, D., Verstege, E.J.M., Groenen, M.A.M., Davies, W., Hoyheim, B., Keiserud, A., Andersson, L., Ellegren, H., Johansson, M., Marklund, L., Miller, J.R., Dear, D.V.A., Signer, E., Jeffreys, A.J., Moran, C., Letissier, P., Muladno, Rothschild, M.F., Tuggle, C.K., Vaske, D., Helm, J., Liu, H.C., Rahman, A., Yu, T.P., Larson, R.G. & Schmitz, C.B., 1995. The PigMap consortium linkage map of the pig (Sus scrofa). Mamm. Genome 6, 157-175. [ Links ]
Chang, Q., Zhou, K.Y., Wang, Y.Q., Zhang, Z.K. & Cao, X., 1999. RAPD analysis of genetic diversity and phylogenetic relationship of the Taihu pig. Acta Genetica Sinica 26, 480-488. [ Links ]
Chen, Y.Z., 1994. Genotyping and application of VNTRs in pigs. Verlag Ulrich. Grauer, Stuttgart, Germany. [ Links ]
Ducos, A., Bidanel, J.P. & Naveau, J., 1992. Estimation of genetic-parameters and genetic trends for production traits in the Sino-European tiameslan composite line. J. Anim. Breed. Genet. 109, 108-118. [ Links ]
Fan, B., Wang, Z.G., Li, Y.J., Zhao, X.L., Liu, B., Zhao, S.H., Yu, M., Li, M.H., Chen, S.L., Xiong, T.A. & Li, K., 2002. Genetic variation analysis within and among Chinese indigenous swine populations using microsatellite markers. Anim. Genet. 33, 422-427. [ Links ]
Fujii, J., Otsu, K., Zorzato, F., Deleon, S., Khanna, V.K., Weiler, J.E., Obrien, P.J. & Maclennan, D.H., 1991. Identification of a mutation in porcine rynaodine receptor associated with malignant hyperthermia. Science 253, 448-451. [ Links ]
Geldermann, H., Muller, E., Beeckmann, P., Knorr, C., Yue, G. & Moser, G., 1996. Mapping of quantitative-trait loci by means of marker genes in F-2 generations of wild boar, Pietrain and Meishan pigs. J. Anim. Breed. Genet. 113, 381-387. [ Links ]
Giuffra, E., Kijas, J.M.H., Amarger, V., Carlborg, O., Jeon, J.T. & Andersson, L., 2000. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics 154, 1785-1791. [ Links ]
Goldstein, D.B., Linares, A.R., Cavallisforza, L.L. & Feldman, M.W., 1995. Genetic absolute dating based on microsatellites and the origin of modern humans. Pro. Natl. Acad. Sci. (USA) 92, 6723-6727. [ Links ]
Goldstein, D.B. & Pollock, D.D., 1997. Launching microsatellites: A review of mutation processes and methods of phylogenetic inference. J. Heredity 88, 335-342. [ Links ]
Grobet, L., Hanset, R. & Dasnois, C., 1992. Halothane challenge test and genotype at the ryanodine receptor locus (Ryr1) in crossbred Pietrain pigs. Ann. Med. Vet. 136, 249-257. [ Links ]
Houde, A., Pommier, S.A. & Roy, R., 1993. Detection of the ryanodine receptor mutation associated with malignant hyperthermia in purebred swine populations. J. Anim. Sci. 71, 1414-1418. [ Links ]
Jiang, Z., Rottmann, O.J., Krebs, O., Chen, J., Liu, H. & Pirchner, F., 2002. A missense mutation in the follicle stimulating hormone receptor (FSHR) gene shows different allele effects on litter size in Chinese Erhualian and German Landrace pigs. J. Anim. Breed. Genet. 119, 335-341. [ Links ]
Kim, K.S. &Choi, C.B., 2002. Genetic structure of Korean native pig using microsatellite markers. Korean J. Genet. 24, 1-7. [ Links ]
Knorr, C., Schwille, M., Moser, G., Muller, E., Bartenschlager, H. & Geldermann, H., 1994. Calcium-release-channel genotypes in several pig-populations - Associations with halothane and Ck reactions. J. Anim. Breed. Genet. 111, 243-252. [ Links ]
Kumar, S., Tamura, K., Jakobsen, I.B. & Nei, M., 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Arizona State University, Tempe, Arizona, USA. [ Links ]
Laval, G., Iannuccelli, N., Legault, C., Milan, D., Groenen, M.A.M., Giuffra, E., Andersson, L., Nissen, P.H., Jorgensen, C.B., Beeckmann, P., Geldermann, H., Foulley, J.L., Chevalet, C. & Ollivier, L., 2000. Genetic diversity of eleven European pig breeds. Genet. Sel. Evol. 32, 187-203. [ Links ]
Lemus-Flores, C., Ulloa-Arvizu, R., Ramos-Kuri, M., Estrada, F.J. & Alonso, R.A., 2001. Genetic analysis of Mexican hairless pig populations. J. Anim. Sci. 79, 3021-3026. [ Links ]
Lewis, P.O. & Zaykin, D., 2000. Genetic Data Analysis: Computer program for the analysis of allelic data. Version 1.0. http://alleyn.eeb.uconn.edu/gda. [ Links ]
Li, K., Li, X., Fan, B., Gong, Y., Zhao, S., Peng, Z. & Liu, B., 2000. The genetic diversity of seven pig breeds in China evaluated by means of microsatellites. Asian Aust. J. Anim. Sci. 13, 100-102. [ Links ]
Li, M.D. &Enfield, F.D., 1989. A characterization of Chinese breeds of swine using cluster-analysis. J. Anim. Breed. Genet. 106, 379-388. [ Links ]
Miller, S.A., Dykes, D.D. & Polesky, H.F., 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215-1215. [ Links ]
Minch, E., 1996. Microsat, the microsatellite distance program. http://human.stanford.edu/microsat/microsat.html. [ Links ]
Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583-590. [ Links ]
Paszek, A., Flickinger, G.H., Fontanesi, L., Beattie, C.W., Rohrer, G.A., Alexander, L. & Schook, L.B., 1998. Evaluating evolutionary divergence with microsatellites. J. Mol. Evol. 46, 121-126. [ Links ]
Rohrer, G.A., Alexander, L.J., Hu, Z.L., Smith, T.P.L., Keele, J.W. & Beattie, C.W., 1996. A comprehensive map of the porcine genome. Genome Res. 6, 371-391. [ Links ]
Rothschild, M.F. & Ruvinsky, A., 1998. Genetics in pig. CAB International. University Press, Cambridge. pp. 39-45. [ Links ]
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G., 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876-4882. [ Links ]
Yue, G.H., 1994. Reproductive performance of Chinese Erhualian pigs. Wrld Rev. Anim. Prod. 3-4, 73-78. [ Links ]
Yue, G.H., Beeckmann, P., Bartenschlager, H., Moser, G. & Geldermann, H., 1999. Rapid and precise genotyping of porcine microsatellites. Electrophoresis 20, 3358-3363. [ Links ]
Yue, G.H., Beeckmann, P., Gerhard, M., Muller, E., Bartenschlager, H., Cepica, S., Schroffel, J., Stratil, A. & Geldermann, H., 2003. QTL alleles on chromosome 7 from fatty Meishan pigs reduce fat deposition. Science in China Series C 46, 10-17. [ Links ]
Yue, G.H., Chen, F. & Orban, L., 2000. Rapid isolation and characterization of microsatellites from the genome of Asian arowana (Scleropages formosus, Osteoglossidae, Pisces). Mol. Ecol. 9, 1007-1009. [ Links ]
Zhang, Z., 1986. Pig breeds in China. Shanghai Science and Technology Press, Shanghai. [ Links ]
# Corresponding author. E-mail: genhau@tll.org.sg














