Services on Demand
Article
Indicators
Related links
-
 Cited by Google
Cited by Google -
 Similars in Google
Similars in Google
Share
SAMJ: South African Medical Journal
On-line version ISSN 2078-5135
Print version ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.112 n.8 Pretoria Aug. 2022
http://dx.doi.org/10.7196/SAMJ.2022.v112i8.16351
RESEARCH
https://doi.org/10.7196/samj.2022.v112i8.16351
Carbapenem-resistant Enterobacterales in patients with bacteraemia at tertiary academic hospitals in South Africa, 2019 - 2020: An update
M LoweI; L ShupingII; O PerovicIII, IV
IPhD; Centre for Healthcare-associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa
IIMPH; Centre for Healthcare-associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa
IIIMMed (Microbiol), FC Path (SA); Centre for Healthcare-associated Infections, Antimicrobial Resistance and Mycoses, National Institute for Communicable Diseases, National Health Laboratory Service, Johannesburg, South Africa
IVMMed (Microbiol), FC Path (SA); Department of Clinical Microbiology and Infectious Diseases, School of Pathology, Faculty of Health Sciences, University of the Witwatersrand., Johannesburg, South Africa
ABSTRACT
BACKGROUND: The emergence of carbapenem-resistant Enterobacterales (CRE) has become a serious and significant public health threat worldwide, owing to the limited antimicrobial therapy options, and the elevated mortality rates associated with these infections
OBJECTIVES: To present an update on the epidemiology of CRE bloodstream infections among hospitalised patients reported under the Group for Enteric, Respiratory and Meningeal Diseases Surveillance in South Africa (GERMS-SA) between January 2019 and December 2020
METHODS: Patients of all ages with CRE bacteraemia were included and isolates, when available, were sent to the reference laboratory for confirmatory testing and molecular characterisation. Multivariable logistic regression analysis was performed to assess factors associated with in-hospital mortality
RESULTS: We included 2 144 patients with CRE bacteraemia with a median age of 33 (interquartile range 1-51) years, of whom 1 145 (54.2%) were male. Klebsiella pneumoniae accounted for 79.8% of infections (n=863/l 082), of which 89.5% (n=611/683) were healthcare associated (HA). The most common carbapenemase genes were carbapenem-hydrolysing oxacillinase-48 (bla0XA_48-like) (76.8%; n=761/991), New Delhi metallo-β-lactamase (blaNDM) (21.1%; n=209/991) and Verona integron-encoded metallo-β-lactamase (blaVIM) (1.3%; n=13/991). None of the screened isolates with a colistin minimum inhibitory concentration >2 (ig/mL harboured the mobilised colistin resistance (mcr)-1 to mcr-5 genes. The crude in-hospital mortality rate was 36.6% (n=377/1 029). Patients aged >60 years (v. 1.6-9 years) (adjusted odds ratio (aOR) 4.53; 95% confidence interval (CI) 2.21 - 9.28), those with comorbidities (diabetes, malignancy, renal and/or cardiovascular failure) (aOR 1.72; 95% CI 1.17 - 2.52), those with altered mental state (aOR 5.36; 95% CI 3.21 - 8.92) and those with previous antimicrobial use (aOR 1.88; 95% CI 1.27 - 2.77) had increased odds of in-hospital mortality
CONCLUSION: The epidemiology of CRE bloodstream infections remained similar compared with the previous surveillance report. Most infections were HA and caused by OXA-48-like carbapenemase-producing K. pneumoniae with no plasmid-mediated colistin resistance. Standard infection control measures should be strengthened
Enterobacterales are Gram-negative bacteria that cause healthcare-associated (HA) and community-associated (CA) infections.[1,2] With the increased use of carbapenems (i.e. doripenem, ertapenem, imipenem and meropenem), the emergence of carbapenem-resistant Enterobacterales (CRE) has become a serious and significant public health threat worldwide.[1,3] The European Centre for Disease Control and Prevention reported that mortality is >50% in patients with CRE bloodstream infections.[4]
Some Gram-negative bacteria can produce carbapenemases (the most common being Guiana extended-spectrum (β-lactamase (GES), imipenem metallo-β-lactamase (IMP), Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), carbapenem-hydrolysing oxacillinase-48 (OXA-48-like) and Verona integron-encoded metallo-β-lactamase (VIM), and they are known as carbapenemase-producing Enterobacterales (CPE).[5] These carbapenemases are commonly harboured on plasmids and can easily be transferred between the members of the Enterobacterales family[4,6] The first CPE were detected in South Africa (SA) in 2011: the KPC and NDM carbapenemases were detected in 2011, followed by the OXA-48-like, GES and VIM carbapenemases in 2012.[7-10] Since then, there has been an increase in Enterobacterales producing one or more carbapenemases.[11] NDM was the most frequently detected carbapenemase in SA from 2011 to 2015.[11-13] However, towards the end of 2015, there was a substantial increase of Klebsiella pneumoniae with OXA-48-like variants across north-eastern SA,[14] and this is currently the most frequently detected carbapenemase in the country, as well as in other regions of the world.[5,8,13,15-19] Routine surveillance for monitoring CRE trends in SA hospitals has been conducted since 2015, and findings from this surveillance have been published previously[13]
We present an updated overview of CRE infections in selected public hospitals in SA from January 2019 to December 2020.
Methods
Study setting and population
This was a cross-sectional study of patients with CRE bacteraemia reported to the Group for Enteric, Respiratory and Meningeal Diseases Surveillance in South Africa (GERMS-SA) from January 2019 to December 2020. Surveillance methods for the programme are detailed elsewhere.[13] Briefly, the study sites were 16 public sector tertiary academic hospitals across four provinces of SA (Gauteng (n=4), KwaZulu-Natal (n=7), Western Cape (n=3), and Free State (n=2)). All patients with CRE bloodstream infections were included in the surveillance programme, and isolates, when available, were sent to the reference laboratory at the National Institute for Communicable Diseases (NICD) for confirmatory testing. Systematic and independent examinations (i.e. audits) of the diagnostic laboratories' records were done to ensure that all cases of CRE bloodstream infection were reported. Surveillance officers collected demographic and clinical information on patients through medical record reviews and patient/caretaker interviews using standard case report forms (CRFs). As patients were not followed up, the in-hospital outcome was documented at the time of CRE completion.
A case of CRE bacteraemia was defined as any patient with Enterobacterales cultured from blood that was resistant to one or more carbapenem (doripenem, ertapenem, imipenem and/or meropenem) or had a positive result for the modified Hodge test according to the Clinical Laboratory Standards Institute (CLSI) guidelines.[20,21] When a patient had an additional Enterobacterales isolated >21 days after the first confirmed laboratory diagnosis, it was regarded as a new case. HA CRE bacteraemia was defined as collection of a positive CRE blood culture specimen >3 days after hospital admission or when a patient had any healthcare contact within 1 year before the current admission. CA CRE bacteraemia was defined as the collection of a positive CRE blood specimen within 2 days of hospital admission when there had been no prior healthcare contact. The source of infection was determined by the surveillance officers based on clinical assessment and/or medical records. Underlying conditions were considered as an acute or chronic comorbidity or condition not related to sepsis and included diabetes mellitus, malignancy, renal failure, cardiovascular diseases, hypertension and tuberculosis. HIV status was reported separately.
Phenotypic and molecular characterisation
Among isolates received at the reference laboratory, viable isolates were identified using an automated system, Microflex matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF) (Bruker Daltonik GmbH, Germany). Antimicrobial susceptibility testing was performed using the MicroScan Walkaway system (Siemens Healthcare Diagnostics Inc., USA) with the NM44 card (Beckman Coulter Inc., USA). All colistin-resistant isolates (identified with the MicroScan Walkaway system (Siemens Healthcare Diagnostics Inc., USA)) were re-tested and confirmed with the Sensititre instrument (Trek Diagnostic Systems Ltd, UK) using the FRCOL panel (Separation Scientific SA (Pty) Ltd, SA). The minimum inhibitory concentration (MIC) results were interpreted using the 2020 CLSI guidelines.[21]
DNA was extracted from all CRE isolates using a crude boiling method.[16] The extracted DNA was used to screen for carbapenemase genes (blaGES (GES-1 - 9 and 11), blaIMP(IMP-9, 16, 18, 22 and 25), blaKPC, blaNDM-1 blaOXA-48-like (i.e. OXA-162, 163, 181, 204, 232, 244, 245 and 247) and blaVIM (VIM-1 - 36)) by a multiplex real-time polymerase chain reaction (PCR) assay (LightCycler 480 II; Roche Diagnostics Corp., USA).[16] Colistin-resistant CRE isolates (with an MIC >2 μg/mL) were screened for the presence of mobilised colistin resistance (mcr)-l to mcr-5 genes by a conventional multiplex PCR.[22]
Data analysis
We conducted a descriptive analysis of demographic and clinical characteristics of all cases of CRE bacteraemia with available data. Data were summarised using percentages and medians with interquartile ranges (IQRs). We conducted univariate and multivariable logistic regression analysis to explore factors associated with in-hospital mortality. We selected variables a priori based on their likelihood to contribute to death (e.g. HIV infection, comorbidities, and prior systemic antibiotics (as a proxy for prior infection/s)) and indicator variables for the severity of illness (e.g. mental status and intensive care unit admission). HIV status was included as one of the covariates possibly contributing to mortality, but this information was not available for patients aged <18 months; this analysis was therefore limited to patients aged > 18 months. We included variables in the multivariable model if the p-value for the association was <0.2 in the univariate analysis; sex and age groups were included in the final model regardless of the p-value. All statistical analyses were done using the Stata statistical software package, version 14 (StataCorp, USA).
Results
Overview of patients with CRE bacteraemia
A total of 2 144 patients with CRE bacteraemia were reported during the 2-year surveillance period (Fig. 1). The majority of the patients were reported from surveillance sites in Gauteng province (64.6%; n=1385/2144), followed by KwaZulu-Natal (19.6%; n=420/2144), Western Cape (13.9%; n=298/2144), and Free State (1.9%; n=41/2144) provinces.
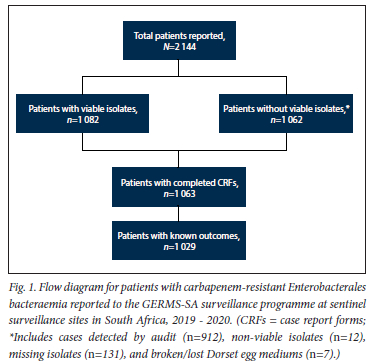
Clinical characteristics of patients with CRE bacteraemia
The median (IQR) age was 33 (1 - 51) years. Male patients accounted for 54.2% (n=l 145/2 113) of the patients with known sex (Table 1). Of the 2 144 reported patients, 49.6% (n=l 063) had completed CRFs and 89.5% (611/683) had HA infections. The proportion representing CA infections in the current report was 10.5% (n=72) v. 4.8% (n=75/l 554) in the previous report[13] (p<0.001). Of those with available information on underlying conditions, 52.3% (n=489/935) had at least one. Ninety-eight percent (n=961/985) had an invasive device inserted at any point from admission to positive specimen collection. Of the 597 patients with HIV status available, 31.0% (n=185) were HIV positive. The majority of the patients (61.0%: n=569/933) had received systemic antibiotics in the past 6 months. Outcome data were available for 1 029 patients, and the in-hospital mortality rate was 36.6% (n=377). This mortality rate was similar to the previous period[13] (37.8%; n=489/l 239) (p=0.559). Seven hundred and fifty-two patients (73.1%) with outcome data also had outcome date available: the median (IQR) duration from specimen collection to outcome was 8 (4 - 17) days, that for patients who were still admitted was 8 (6 - 14) days, that for discharged patients was 17 (8 - 32) days, and that for patients who died was 4(1-10) days.
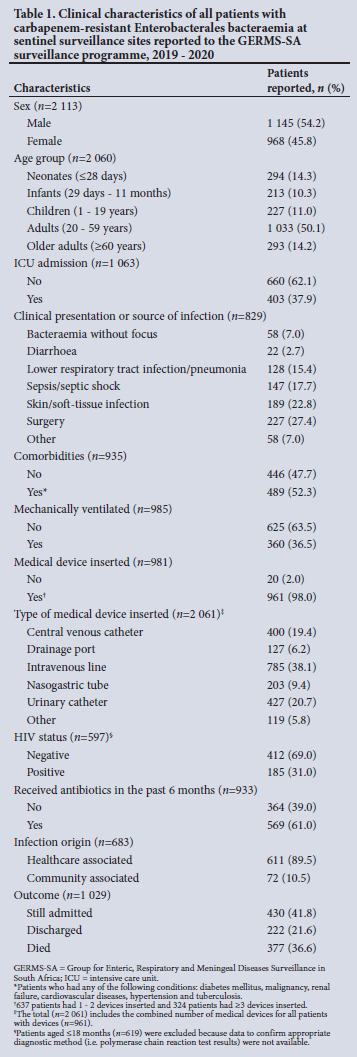
Mortality among patients with CRE bacteraemia aged >18 months
Of the 1 552 patients aged >18 months, 763 (49.2%) had outcome data, and 306 (40.1%) of these patients died. A higher proportion of females compared with males died (41.5% v. 38.9%, respectively), and of adults aged >60 years compared with children aged 1.6 -19 years (56.8% v. 22.2%, respectively) (Table 2). In the multivariable logistic regression model, patients aged >60 years (adjusted odds ratio (aOR) 4.53; 95% confidence interval (CI) 2.21 - 9.28;p<0.001), those with other comorbidities (such as diabetes, malignancy renal and/or cardiovascular failure) (aOR 1.72; 95% CI 1.17 - 2.52: p=0.006), those with altered mental state (aOR 5.36; 95% CI 3.21 -8.92; p<0.001), and those with previous antimicrobial use (aOR 1.88; 95% CI 1.27 - 2.77; p=0.001) had increased odds of in-hospital mortality.
CRE isolates
Of all patients, 50.5% (n=1082/2144) had at least one viable isolate and a confirmatory species identity (Fig. 1). Overall, K. pneumoniae (79.8%; n=863/1082) accounted for the majority of infections, followed by Enterobacter cloacae complex (5.7%; n=62/1082), Serratia marcescens (5.0%; n=54/1082), and Escherichia coli (4.1%; n=44/1082). The proportion of Enterobacterales isolated was similar in 2019 and 2020 (Fig. 2).
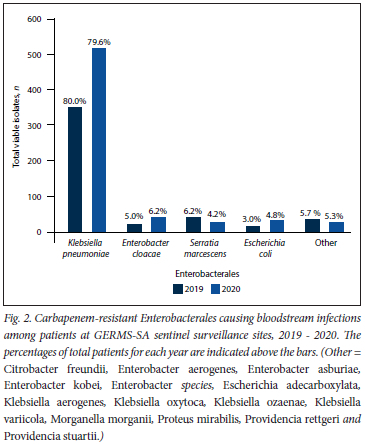
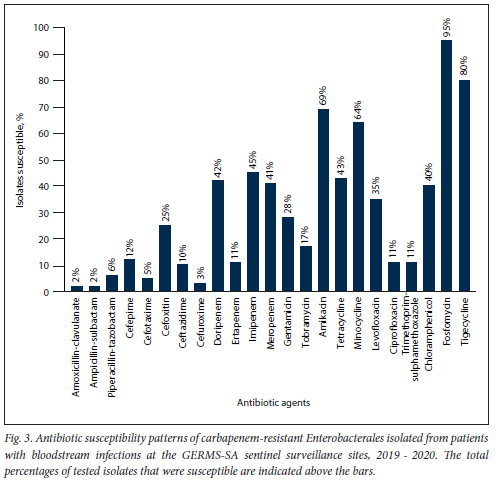
Susceptibility to a majority of the antibiotics tested was <30% (Fig. 3). Susceptibility to amikacin (69.4%; n=751/1082), fosfomycin (95.4%; n=1 032/1082), minocycline (64.4%; n=697/l 082) and tigecycline (80.2%; n=868/1082) was >60%. Colistin resistance was reported in 18.6% (n=201/1079) of the isolates, and 81.4% (n=878/l 079) of the isolates had intermediate resistance. Of the 201 colistin-resistant isolates, 52 (i.e. S. marcescens (n=49), Morganella morganii (n=2) and P.roteus mirabilis (n=1)) were intrinsically resistant (25.9%). The proportion of isolates susceptible to doripenem, imipenem and meropenem ranged from 41.2% to 44.9%, while susceptibility to ertapenem was 11.5% (n=124/1082).
The MIC50 and MIC90 (MIC required to inhibit the growth of 50.0% and 90% of organisms, respectively) were the same for doripenem (both >4 μg/mL), ertapenem (both >1 μg/mL) and meropenem (both 8 ug/mL), while for imipenem they were 2 μg/mL and 8 μg/mL, respectively. The MIC distribution for all carbapenems is shown in Table 3.
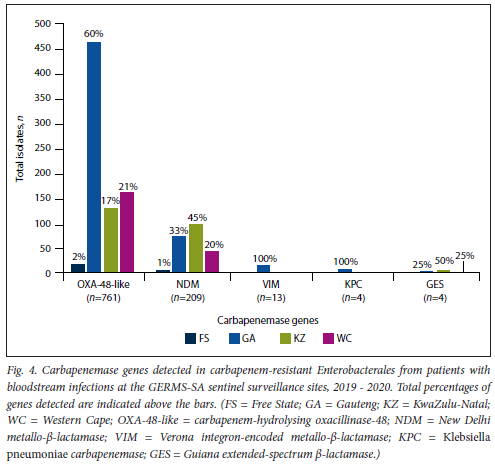
Of the 1082 patients with viable isolates, 915 (84.6%) had isolates with one carbapene-mase gene detected, 38 (3.5%) had isolates with two genes detected and 129 (11.9%) had isolates with no genes detected. All the Escherichia adecarboxylata, M. morganii and P. mirabilis isolates tested negative for all the screened carbapenemase genes. Overall, the most common carbapenemases were blaoxA-48-like (76.8%; n=761/991), followed by blaNDM (21.1%; n=209/991), blaVIM (1.3%; n=13/991), blaGES (0.4%; 4/991) and blaKPC (0.4%; n=4/991) (Fig. 4). The blaOXA-48-like gene was predominant in all four provinces, although the proportion of blaNDM in KwaZulu-Natal was notably high (42%; n=95/224). The blaIMP gene was not detected during this surveillance period. Among the isolates that harboured two carbapenemase genes simultaneously, the combinations were blaoxA-48-iike with blaNDM (71.1%; 11=27/38), blaoxA-48-iike with blaKPC (7.9%; n=3/38), blaoxA-48-iike with blaVIM (7.9%; 3/38), blaoxA-48-iike with blaGES (5.3%; n=2/38), blaNDM with blaGES (5.3%; n=3/38), and blaNDM with blaKPC (2.6%; n=1/38). The screened CRE isolates (n=159) with colistin MIC >2 μg/mL were negative for the mcr-l to mcr-5 genes.
Carbapenem susceptibility was compared with the identified carbapenemase genes (Fig. 5). The majority of the isolates harbouring the blaoxA-48-iike gene (n=761) were resistant to ertapenem (n=576) and meropenem (n=377), while the remaining isolates were susceptible to doripenem (n=353) and imipenem (n=388). The majority of the isolates harbouring the blaNDM gene (n=209) were resistant to doripenem (n=200), ertapenem (n=205), imipenem (n=200) and meropenem (n=198). The majority of the isolates harbouring the blaVIM gene (n=13) were intermediately resistant to doripenem (n=6) and ertapenem (n=6), resistant to imipenem (n=10), and susceptible to meropenem (n=9). The majority of the isolates harbouring the blaGES gene (n=4) were resistant to doripenem (n=3), imipenem (n=3) and meropenem (n=3), while all isolates were resistant to ertapenem (n=4). The majority of the isolates harbouring the blaKPC gene (n=4) were resistant to imipenem (n=3), while all isolates were resistant to doripenem (n=4), ertapenem (n=4) and meropenem (n=4).
Discussion
This study showed the current epidemiology of CRE bloodstream infections among hospitalised patients with CRE bacteraemia in SA. Similar to the previous period, most CRE infections occurred in Gauteng, KwaZulu-Natal and Western Cape.[13] However, the actual proportion of patients from Gauteng decreased, while it increased in Western Cape and KwaZulu-Natal. The increase was most notable in Western Cape, where the proportion of patients doubled. This shift may be due to changes in at-risk patients being admitted and/or specimen-taking practices, an increase in the number of surveillance sites, and increasing or decreasing numbers of patients admitted. These data were not collected as part of the surveillance, so the reasons for the changes are unclear. However, the observed increase in patients in some provinces is of concern and should be closely monitored.
The in-hospital mortality rate was high, but similar to the previous surveillance period.[13] Zou et al.[23] reported a higher mortality rate (50%; n=40/80) among hospitalised patients with CRE infections in China. However, a study conducted across 15 states in the USA[3] showed a lower in-hospital mortality rate in patients with a CRE infection (24%; n=107/449) compared with our findings. Our study showed that patients aged >60 years, those with other comorbidities, those with altered mental state, and those with previous antimicrobial exposure had increased odds of in-hospital mortality, which is in agreement with previous findings.[24] Our findings show consistently high mortality in patients with CRE infections and highlight patients at increased risk of death who may benefit from intensified prevention measures, such as proper and timely clinical management and treatment, active screening on admission to hospital or a specific high-risk ward, and pre-emptive contact precautions to reduce infections and consequently CRE-related mortality[25]
K. pneumoniae accounted for nearly 80% of CRE infections, similar to the previous reporting period.[13] Globally, K. pneumoniae remains the dominant pathogen among patients with CRE infections; it was the most dominant pathogen reported in 33 European countries, as well as in 15 states in the USA and 25 provinces and municipalities in China.[3,17,26,27]
Overall resistance to antibiotics remained high, but the proportion of isolates resistant to tigecycline and fosfomycin decreased slightly compared with the previous SA surveillance report.[13] More than 80% of the isolates had intermediate resistance to colistin. The change from susceptible to intermediate resistance was due to the changes in the 2020 CLSI guidelines (i.e. the susceptible category was removed from the guidelines; only the intermediate (< 2 μg/mL) and resistant (> 4 μg/mL) categories remain). The colistin MIC breakpoints differ in the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guideline (i.e. only the susceptible (<2 μg/mL) and resistant categories (>2 μg/mL) are reported). Compared with the previous surveillance report, colistin resistance has increased by 5.6% (the MIC breakpoint for the resistant category remained the same).[13,21] The high level of carbapenem resistance, especially towards ertapenem, is well described in other SA studies.[11,13,28] This increase in resistance could be due to suboptimal infection prevention and control (IPC) measures and antibiotic stewardship practices. Compared with the previous surveillance report, the MIC50 has stayed in the same reported range for ertapenem (>1 ug/mL) but has increased for doripenem (<1 μg/mL to >4 μg/mL), imipenem (<1 μg/mL to 2 μg/mL) and meropenem (<1 μg/mL to 8 μg/mL), and the MIC90 also stayed in the same reported range for doripenem (>4 ug/mL), ertapenem (>1 ug/mL), imipenem (>4 μg/mL), and meropenem (>4 μg/mL).[13]
The blaOXA-48-like,blaNDM and blaVIM remained the most common carbapenemase genes identified in public sector hospitals in SA overall, but their proportions have changed. Compared with the previous surveillance period, the number of CREs harbouring the blaOXA-48-likeand blaKPC genes has increasedfrom 52% to 76.8% and 0.1 % to 0.4%, respectively. In contrast, the blaNDM gene decreased from 34% to 21.1% and the blaVIM gene from 4% to 1.3%, while the blaIMP gene was not detected compared with being present in 0.2% of the isolates previously. The number of CREs harbouring the blaGES gene remained the same. The increase in the number of CREs harbouring the blaOXA-48-like gene could be due to the fact that plasmids containing this gene can disseminate between various bacterial species via horizontal transmission more efficiently[17] The current spread of the blaOXA-48-like gene is predominantly driven by the composite transposon Tn 1999 and its variants (i.e. Tn 1999.2 and Tn 1999.3), which are harboured on the pOXA-48a-like IncL conjugative plasmid.[17,29] The pOXA-48a-like IncL plasmid is highly transmissible and has a transfer frequency 50 times higher than the pNDM-OM IncL plasmid that harbours the blaNDM-1 gene.[17] IPC measures such as early identification of CPE, contact precautions, hand hygiene, environmental cleaning, adhering to aseptic techniques and antimicrobial stewardship should be implemented, and CRE- and/or CPE-positive patients should ideally be isolated and treated according to strict standard guidelines to curb transmission.[25,30] Screening and isolation may not always be feasible owing to limited resources and staff. Awareness of the specific carbapenemase detected is critical in decisions around optimal antibiotic treatment.[25] CPE is associated with increased mortality and poor clinical outcomes, and is considered to be more virulent than CRE isolates that do not harbour carbapenemase genes.'251 The majority of isolates harbouring the blaGES, blaKPC and blaNDM genes were resistant to all carbapenems. The majority of isolates harbouring the blaVIM gene were resistant to imipenem, intermediately resistant to doripenem and ertapenem, and susceptible to meropenem. The majority of isolates harbouring the blaOXA-48-like gene were resistant to ertapenem and meropenem, susceptible to imipenem, and showed an equal susceptible and resistant distribution to doripenem, which corresponds with previously reported SA results.[11,28]
Just over a tenth of the CRE isolates did not harbour any carbapenemase genes, which was similar to the previous period.[13] CRE isolates that do not harbour carbapenemase genes are generally still resistant to multiple antibiotics and can be transmitted between patients. Their detection therefore still warrants targeted infection control interventions, such as contact precautions.[25]
This study has some limitations: (i) comprehensive patient demographic details were unavailable, and it was not possible to determine accurate trends in patient age, ward and gender; (ii) treatment was not included as a factor associated with mortality in the analysis; (iii) data originated from public sector hospitals in SA; (iv) owing to missing isolates, it was not possible to determine accurate trends in antibiotic resistance and circulating carbapenemase genes; (v) non-carbapenemase-producing Enterobacterales were not screened for other carbapenemases, AmpC and/or extended-spectrum beta-lactamases by PCR; and (vi) the conventional PCR assay used in this study to screen for the presence of mcr genes only covered the mcr-1 to mcr-5 genes; we did not screen for the mcr-6 to mcr-9 genes.
Conclusion
The study findings show overall consistent epidemiology of CRE bloodstream infections with slight changes that may become prominent over time. K. pneumoniae harbouring the blaOXA-48-like gene remains the most prevalent organism among patients with CRE bacteraemia in SAs public academic hospitals. The findings in this study call for better and improved surveillance programmes nationwide.
Declaration. None.
Acknowledgements. The authors thank all staff members of the Antimicrobial Resistance Laboratory and Culture Collection (AMRL-CC), specifically Marshagne Smith, Gloria Molaba, Naseema Bulbulia. Rosah Kganakga, Rubeina Badat and Agnes Sesoko for phenotypic testing, Boniwe Makwakwa for data capturing, and Ruth Mogokotleng, Wilhelmina Strasheim, Nokuthula Linda and Dineo Bogoshi for molecular testing. The authors also thank the GERMS-SA site investigators and surveillance officers.
Author contributions. Surveillance for CRE was initiated by OP. All authors contributed to the study conception and design. Data analysis was performed by ML and LS. ML wrote the first draft and revised it based on feedback from both the co-authors. All authors reviewed and approved the final manuscript.
Funding. This study was supported by the Centre for Healthcare-Associated Infections, Antimicrobial Resistance and Mycoses (CHARM). NICD. No additional funds, grants, or other support were received during the preparation of this manuscript.
Conflicts of interest. None.
References
1. Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by carbapenem-resistant Enterobacteriaceae. An update on therapeutic options. Front Microbiol 2019;10:80. https://doi.org/10.3389/fmicb.2019.00080 [ Links ]
2. Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae. Here is the storm Trends Mol Med 2012;18(5):263-272. https://doi.org/10.1016/j.molmed.2012.03.003 [ Links ]
3. Van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2). A prospective cohort study Lancet Infect Dis 2020(6);20:731-741. https://doi.org/10.1016/S1473-3099(19)30755-8 [ Links ]
4. World Health Organization. Guidelines for the prevention and control of car bap en em-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. Geneva. WHO, 2017. http://apps.who.int/iris/bitstream/handle/10665/259462/9789241550178-eng.pdf,jsessionid=623130C9C3DE356707C020ED2F5D2CC0?sequence=l (accessed 8 June 2021). [ Links ]
5. Castanheira M, Doyle TB, Collingsworth TD, Sader HS, Mendes RE. Increasing frequency of OXA-4 8-producing Enter ob acter ales worldwide and activity of ceftazidime/avibactam, meropenem/ vaborbactam and comparators against these isolates. J Antimicrob Chemother 2021;76(12):3125-3134. https://doi.org/10.1093/jac/dkab306 [ Links ]
6. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 2015;59(10):5873-5884. https://doi.org/10.1128/AAC.01019-15 [ Links ]
7. Brink A, Coetzee J, Clay C, et ai. The spread of car bapenem-resist ant Enter obacteriaceae in South Africa. Risk factors for acquisition and prevention. S Afr Med J 2012;102(7):599-601. https://doi.org/10.7196/SAMJ.5789 [ Links ]
8. Brink AJ, Coetzee J, Clay CG, et aL Emergence of New Delhi metalio-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J Clin Microbiol 2012;50(2):525-527. https://doi.org/10.1128/JCM.05956-11 [ Links ]
9. Brink AJ, Coetzee J, Corcoran C, et al. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacter!aceae in South Africa and evidence of in vivo selection of Colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol 2013;51(1):369-372. https://doi.org/10.1128/JCM.02234-12 [ Links ]
10. Lowman W, Sriruttan C, Nana T. NDM-1 has arrived. First report of carbapenem resistance mechanism in South Africa. S Afr Med J 2011;101(12):873-875. https://doi.org/10.7196/SAMJ.5329 [ Links ]
11. Perovic O, Britz E, Chetty V, Singh-Moodley A. Molecular detection of carbapenemase-producing genes in referral Enter obacteriaceae in South Africa. A short report. S Afr Med J 2016;106(10):975-977. https://doi.org/10.7196/SAMJ.2016.vl06il0.11300 [ Links ]
12. Singh-Moodley A, Perovic O. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enter obacteriaceae in South Africa. BMC Infect Dis 2016;16(1):536-546. https://doi.org/10.1186/sl2879-016-1858-7 [ Links ]
13. Perovic O, Ismail H, Quan V, et al. Carbapenem-resistant Enterobacteriaceae in patients with bacteraemia at tertiary hospitals in South Africa, 2015 to 2018. Eur J Clin Microbiol Infect Dis 2020;39(7):1287-1294. https://doi.org/10.1007/sl0096-020-03845-4 [ Links ]
14. Lowe M, Kock MM, Coetzee J, et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014 - 2016. Emerg Infect Dis 2019;25(4):739-747. https://doi.org/103201/eid2504.181482 [ Links ]
15. Strydom KA, Chen L, Kock MM, et aL Klebsiella pneumoniae ST307 with OXA-181. Threat of a high-risk clone and promiscuous plasmid in a resource-constrained healthcare setting. J Antimicrob Chemother 2020;75(4):896-902. https://doi.org/10.1093/jac/dkz550 [ Links ]
16. Singh-Moodley A, Perovic O. Phenotypic and genotypic correlation of carbapenememase-producing Enterobacteriaceae and problems experienced in routine screening. S Afr Med J 2018;108(6):495-501. https://doi.org/10.7196/SAMJ.2018.v108i6.12878 [ Links ]
17. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev 2019;33(l):e00102-19. https://doi.org/10.1128/CMR.00102-19 [ Links ]
18. Tangden T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: Clinical perspectives on detection, treatment and infection control. J Intern Med 2015;277(5):501-512.https://doi.org/10.1111/joim.12342 [ Links ]
19. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae. Epidemiology, genetic context, treatment options and detection methods. Front Microbiol 2016;7:895. https://doi.org/10.3389/fmicb.2016.00895 [ Links ]
20. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing 29th ed. CLSI supplement M100. Wayne, Pa.. CLSI, 2019. [ Links ]
21. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, Pa.. CLSI, 2020. [ Links ]
22. Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-l, mcr-2, mcr-3, mcr-A and mcr-5 for surveillance purposes. Euro Surveill 2018;23(6):17-00672. https://doi.org/10.2807/1560-7917.ES.2018.23.6.17-00672 [ Links ]
23. Zou H, Xiong SJ, Lin QX, Wu ML, Niu SQ, Huang SF. CP-CRE/non-CP-CRE stratification and CRE resistance mechanism determination help in better managing CRE bacteremia using ceftazidime-avibactam and aztreonam-avibactam. Infect Drug Resist 2019;12:3017-3027. https://doi.org/10.2147/IDR.S219635 [ Links ]
24. Tamma PD, Goodman KE, Harris AD, et ai. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae bacteremia. Clin Infect Dis 2017;64(3):257-264. https://doi.org/10.1093/cid/ciw741 [ Links ]
25. Ambretti S, Bassetti M, Clerici P, et al. Screening for carriage of carbapenem-resistant Enterobacteriaceae in settings of high endemicity. A position paper from an Italian working group on CRE infections. Antimicrob Resist Infect Control 2019;8:136. https://doi.org/10.1186/sl3756-019-0591-6 [ Links ]
26. Glasner C, Albiger B, Buist G, et al. Carbapenemase-producing Enterobacteriaceae in Europe. A survey among national experts from 39 countries, February 2013. Euro Surveill 2013;18(28):20525. https://doi.org/10.2807/1560-7917.es2013.18.28.20525 [ Links ]
27. ZhangR, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 2017;19:98-106. https://doi.org/10.1016/j.ebiom.2017.04.032 [ Links ]
28. Group for Enteric, Respiratory and Meningeal Diseases Surveillance in South Africa (GERMS-SA). Annual surveillance report. 2020. https://www.nicd.acza/wp-content/uploads/2022/02/2020-GERMS-SA-Annual-Review.pdf (accessed 1 December 2021). [ Links ]
29. Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 2012;56(1):359-562. https://doi.org/10.1128/AAC.05289-11 [ Links ]
30. National Department of Health, South Africa. Practical manual for implementation of the national infection prevention and control strategic framework. October 2021. https://wwwknowledgehub.org.za/system/files/elibdowmoads/2021-10/National%20Infection%20Prevention%20and%20Control%20Practical%20Manual%20%28Web%20version%29.pdf (accessed 1 December 2021). [ Links ]
 Correspondence:
Correspondence:
O Perovic
olgap@nicd.ac.za
Accepted 20 April 2022














