Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
SAMJ: South African Medical Journal
versão On-line ISSN 2078-5135
versão impressa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.112 no.2 Pretoria Fev. 2022
http://dx.doi.org/10.7196/SAMJ.2022.v112i2.16052
IN PRACTICE
CASE REPORT
Prolonged SARS-CoV-2 RNA shedding in a young man recovering from traumatic pneumothorax
C N UmunnakweI, II; Z N MakatiniIII, IV; A MdunyelwaV; M MaphangaV; M NijhuisVI, VII, VIII; A WensingIX, X, XI; H A TempelmanIX, X, XII
IBSc, PhD; Ndlovu Laboratories, Elandsdoorn, Limpopo Province, South Africa
IIBSc, PhD; Ndlovu Research Consortium, Elandsdoorn, Limpopo Province, South Africa
IIIMB ChB; Ndlovu Research Consortium, Elandsdoorn, Limpopo Province, South Africa
IVMB ChB; Department of Virology, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
VMSc;Ndlovu Laboratories, Elandsdoorn, Limpopo Province, South Africa
VIPhD; Ndlovu Research Consortium, Elandsdoorn, Limpopo Province, South Africa
VIIPhD; Department of Medical Microbiology, Virology, University Medical Center Utrecht, Netherlands
VIIIPhD; HIV Pathogenesis Research Unit, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
IXMD, PhD; Ndlovu Research Consortium, Elandsdoorn, Limpopo Province, South Africa
XMD, PhD; Department of Medical Microbiology, Virology, University Medical Center Utrecht, Netherlands
XIMD, PhD; Wits Reproductive Health and HIV Institute, University of the Witwatersrand, Johannesburg, South Africa
XIIMD, PhD; Ndlovu Laboratories, Elandsdoorn, Limpopo Province, South Africa
ABSTRACT
We describe a case of prolonged SARS-CoV-2 RNA shedding in an HIV-negative 21-year-old man recovering from abdominal and thoracic trauma. Nasopharyngeal (NP) swabs collected at 12 time points over a 95-day span all tested positive for SARS-CoV-2 by reverse transcription polymerase chain reaction (RT-PCR). Genotyping revealed canonical beta-variant E484K and N501Y mutations at earlier time points. Human rhinovirus, coronavirus NL63 and respiratory syncytial virus B were detected at different time points by RT-PCR. Full blood analysis at time point 9 (day 82) showed leukopenia with lymphocytosis. The patient's NP swab tested negative for SARS-CoV-2 by RT-PCR 101 days after the first positive test. The prolonged duration of SARS-CoV-2 RNA shedding in the context of trauma presented here is unique and has important implications for COVID-19 diagnosis, management and policy guidelines.
RNA shedding and viral load kinetics of SARS-CoV-2, the causative agent of COVID-19, remain incompletely understood. The duration of RNA shedding in respiratory samples has been reported as a median of ~18 days, but shedding is heterogeneous and can occur up to 92 days after symptom onset, in what can be termed prolonged RNA shedding.[1-9] A concise definition of prolonged SARS-CoV-2 RNA shedding remains lacking in the current literature. Some authors define it as detectable RNA shedding >14 days after the onset of symptoms or after the first positive real-time reverse transcription polymerase chain reaction (RT-PCR) result, while others use the median duration of viral RNA shedding in study populations as a cutoff above which shedding is considered prolonged[8,10,11] Although inadequately characterised, prolonged SARS-CoV-2 RNA shedding is not uncommon; there are increasing reports of persistent virus shedding in respiratory samples despite symptom resolution, positive tests after two consecutive negative SARS-CoV-2 molecular tests, and shedding in stool despite respiratory samples testing negative[4,5,10-13]
Factors associated with prolonged SARS-CoV-2 RNA shedding include age, gender, disease severity, comorbidities and immunocompromised status[4,8,14-20] It is hypothesised that higher baseline viral loads in older patients lead to prolonged RNA shedding[3,21] Meanwhile, differences in angiotensin-converting enzyme 2 levels between males and females are postulated to contribute to the sex-based difference in disease response as well as to prolonged SARS-CoV-2 RNA shedding, more commonly found in males.[22,23] In respiratory samples, particularly from the lower respiratory tract, prolonged RNA shedding has been associated with persistently high immunoglobulin M titres up to the 9th week after symptom onset, even in patients who received antiviral therapy such as ribavirin and remdesivir.[10,11,15] SARS-CoV-2 RNA shedding in stool during convalescence as well as in patients without gastrointestinal symptoms has also been documented, with anal swabs from paediatric patients testing positive, in one case up to 42 days after the initial positive test.[11,22] In view of the implications that prolonged shedding could have on patient care and follow-up, infection prevention and mitigation measures, and virus persistence and evolution, elucidating infectivity dynamics and viral shedding kinetics in prolonged shedders remains important.
We describe a unique case, identified at Ndlovu Laboratories, of prolonged SARS-CoV-2 RNA shedding in a 21-year-old man recovering from abdominal pathology and traumatic pneumothorax due to a stabbing incident.
Case presentation
Ndlovu Laboratories is a clinical trial research centre and diagnostic laboratory situated in rural Limpopo Province, South Africa. On 2 January 2021, a 21-year-old man, recently discharged from a local general hospital after treatment of a stab wound, was referred to Ndlovu Laboratories by a private cardiothoracic surgeon for SARS-CoV-2 RT-PCR testing prior to evaluation for decortication surgery. RT-PCR testing performed on the patient's nasopharyngeal (NP) swab was positive for SARS-CoV-2, and on subsequent testing, household close contacts, comprising the patient's mother and girlfriend, were also SARS-CoV-2-positive. The patient and close contacts were instructed to undergo isolation for 14 days, and guidance was provided by the surgeon to re-test the patient after isolation.
Upon request and with the patient's consent, medical records were obtained from the hospital where the patient had been admitted. Available medical records indicated admission on 8 December 2020, at which point chest radiographs showed no evidence of a pneumothorax. On the third day of the admission (10 December 2020), chest radiographs revealed an elevated hemidiaphragm that could have been secondary to a pneumoperitoneum from a ruptured stomach or free air trapped under the diaphragm. An explorative laparotomy revealed a laceration in the stomach, which was sutured. The medical records indicated that on return from theatre, the patient had difficulty breathing and was given oxygen. A chest radiograph was repeated on 14 December, and an intercostal (IC) drain was inserted (Fig. 1A). Radiographs were repeated on 15, 17 and 21 December. The radiograph of the 17th revealed evidence of loculated empyema (Fig. 1A, red arrow), raising the possibility of loculation as a result of infection or thoracic trauma. On 21 December, the IC drain was removed and the patient was discharged on analgesics and antibiotics. The medical records also indicated that the patient was briefly readmitted with dyspnoea, and subsequently referred to another general hospital for decortication surgery. At this point, he opted for evaluation by a private cardiothoracic surgeon, who requested a COVID-19 test from Ndlovu Laboratories, resulting in the SARS-CoV-2 RT-PCR test performed on 2 January 2021.
Follow-up
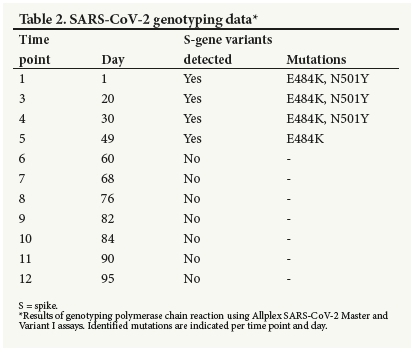
On 16 January, the patient was retested for SARS-CoV-2. The RT-PCR result was positive, and he continued to test positive for 10 more consecutive visits spanning a total of 95 days (2 January -6 April 2021). During this period, apart from difficulty in breathing, reported only during physical exertion, he remained asymptomatic and cycle threshold (Ct) values steadily increased over time, indicating decreasing viral loads (Fig. 2 and Supplementary Table 1, the table available at https://www.samedical.org/file/1771). To assess whether long-term SARS-CoV-2 RNA shedding was detectable in different anatomical sites, SARS-CoV-2 RT-PCR was performed on matched NP, oropharyngeal (OP), saliva, sputum and cheek cell specimens, collected at time point 8 (day 76), and on plasma obtained at time point 9 (day 82). Only the NP sample tested positive for SARS-CoV-2, all the other specimens being negative. To gain insight into persistent SARS-CoV-2 RNA detection, a full blood count (FBC) was also performed at time point 9 (day 82). The FBC showed a decreased white blood cell (WBC) count and elevated lymphocytes suggestive of inflammation (Table 1). HIV-1/2 rapid testing conducted during follow-up was negative. To explore secondary and/or underlying viral/bacterial respiratory infections that could potentially contribute to SARS-CoV-2 persistence, RT-PCR for a comprehensive panel of respiratory pathogens was retrospectively performed on NP swabs from all SARS"CoV"2"positive time points (1 through 12). Results revealed infections with human rhinovirus at time point 5 (day 49), human coronavirus NL63 at time point 10 (day 84), and respiratory syncytial virus B at time points 11 and 12 (days 90 and 95). To assess the likelihood of reinfection and/or infectiousness, using the presence of viral antigen as a proxy, a rapid COVID-19 antigen test was performed at time point 8 (day 76) and the result was negative, suggesting that the subject was not actively infected or infectious at the time. Furthermore, SARS-CoV-2 RT-PCR of close contacts performed at time points 11 and 12 (days 90 and 95) were negative, further supporting that the subject was non-infectious at that time. To explore the likelihood of ongoing SARS-CoV-2 replication, using viral evolution as a proxy, the first seven time-point samples (days 1 - 68) were submitted for Illumina whole-genome sequencing (WGS) (Illunima Inc., USA). However, coverage of the genome in all samples was extremely low (3 - 8%), which can be attributed to relatively high Ct values, especially at later time points. The viral sequence at all 12 positive time points (days 1 - 95) was also examined using SARS-CoV-2 genotyping kits capable of detecting and differentiating between spike gene (S-gene) variants. Results indicated that canonical B.1.531 (beta variant) mutations, E484K and N501Y, were present at the first four time points (days 1 - 30). The E484K but not the N501Y mutation was detected at the fifth time point (day 49) and S-gene variants were not detected at any later time points (Table 2). A summary of all diagnostics performed is provided in Fig. 3 (see also 'Material and methods', supplementary material available at https://www.samedical.org/file/1770).
Outcome and resolution
On 12 April, at the 13th sampling time point and 101 days after the first positive test, the patient tested negative for SARS-CoV-2. A follow-up test a week later on 19 April confirmed his negative status. Five chest radiographs obtained longitudinally during his testing period at Ndlovu Laboratories reflected gradual radiological resolution (Fig. 1B). In particular, the final chest radiograph showed notable healing of the lungs, obviating the need for surgery.
Discussion
SARS-CoV-2 shedding is a heterogeneous phenomenon and differs according to the sampled site, with maximum shedding periods of 83 days in the upper respiratory tract, 59 days in the lower respiratory tract, 126 days in stool, and 60 days in serum having been reported.[4] We describe an unusual case of persistent SARS-CoV-2 RT-PCR positivity in NP samples for at least 95 days in an HIV-negative 21 -year-old man with no underlying comorbidities, albeit recovering from abdominal and lung trauma. Several aspects of this case make it unique. The most striking feature is SARS-CoV-2 infection occurring in the context of significant trauma in the patient. It is likely that this unique combination of infection and extensive pulmonary and abdominal damage contributed significantly to delayed viral clearance. Of note, the patient began experiencing difficulty in breathing only after surgery, and evidence of pneumothorax appeared after an IC drain was inserted, suggesting that clinical management exacerbated trauma, while also raising the possibility of nosocomial SARS-CoV-2 infection.
An additional important feature of the case is the patient's HIV-negative status. Several reports demonstrate that being immunocompromised is a major factor contributing to prolonged SARS-CoV-2 shedding.[24-28] SARS-CoV-2 infection in severely immunocompromised individuals is believed to contribute to emergence of new variants.[19,20] We speculate that the trauma suffered by the patient may have compromised his immune system, increasing susceptibility to SARS-CoV-2 and other respiratory infections and predisposing him to delayed viral clearance. The presence of other respiratory infections detected at different sampling points supports this hypothesis. We would recommend that such individuals should be prioritised for vaccination to minimise the risk of viral persistence and intra-host viral evolution.
Other factors associated with persistent virus shedding include comorbidities, lymphopenia and, more controversially, integration of viral fragments into the host genome.[5,10-13,16,23,29,30] Our patient had no known underlying comorbidities and no history of smoking or drinking. Although his WBC count was low, lymphopenia was not observed; rather, lymphocytosis in the context of SARS-CoV-2 infection was seen, further underlining the uniqueness of the case. In addition, all attempts at amplifying SARS-CoV-2 from the patient's epithelial cheek cell genome yielded negative results.
It should be noted that the patient's persistent RT-PCR-positive results prevented him from being evaluated for surgery and limited his access to care. However, a positive PCR result does not always imply the presence of infectious virus. To distinguish between replicating/infectious virus and non-replicating/non-infectious virus, additional testing such as virus culture is required to establish virus viability. Although virus culture was not performed in this case, we can deduce that at later time points only non-infectious virus was persistently detected, based on high Ct values observed at later time points, failure to obtain coverage of the viral genome through WGS, and negative rapid antigen results. Our data cannot categorically rule out the possibility of very low-level viral replication in specific anatomical sites/compartments, especially as SARS-CoV-2 was only detected in NP specimens and could not be detected in OP, sputum, saliva, serum or cheek cell specimens.
Several systematic reviews have indicated that although virus shedding may be prolonged, the duration of replication-competent and infectious virus is short, and individuals, particularly mildly and moderately infected, have been found to less likely be infectious >10 days after symptom onset.[2,4,31-33] Positive SARS-CoV-2 PCR results should therefore not preclude admittance for surgery; rather, individuals' Ct values and medical history should be reviewed on a case-by-case basis.
Study limitations
Limitations of this study include that baseline measurements for haematological analysis, rapid antigen tests at early sampling time points, and serological analysis were all not performed. The study is an analysis of a single individual, and caution should be exercised in generalising. Genotyping with commercial kits, such as the one used in our study, are limited to detection of specific mutations and cannot provide information on clinically relevant mutations that can be present elsewhere in the viral genome. Finally, medical records obtained from the local hospital where the patient was admitted were incomplete.
Teaching points
This case study demonstrates complementary approaches that can be leveraged to gain important insight into SARS-CoV-2 transmission and viral dynamics, and inform on proper COVID management, prevention and control measures.
Prolonged shedders should be reviewed on a case-by-case basis, taking into account medical history, immunocompromised status, and whether or not persistent positivity stems from infectious virus.
Analysis of Ct values combined with rapid antigen testing can act as a proxy for potential infectiousness of prolonged shedders. If PCR results of prolonged shedders show an increasing trend of Ct values over time, a negative rapid antigen result can rule out infectiousness and reinfection, especially if the last median PCR Ct is >34 (time point 4 in our case study, when all three gene targets are no longer consistently detected; Supplementary Table 1, https://www.samedical.org/file/1771). In such cases, the subject poses virtually no infection risk, and the persistent PCR-positive results should not contraindicate admittance for hospital care, including surgery.
Trauma patients may be at increased risk of nosocomial SARS-CoV-2 infection and should be tested routinely during hospital admission and isolated from potential sources of SARS-CoV-2 infection. Medical procedures that could place such individuals at further risk of SARS-CoV-2 infection should be avoided unless necessary.
Immunocompromised individuals and trauma patients are at increased risk of prolonged viral shedding and should be prioritised for vaccination.
Declaration. None.
Acknowledgements. We thank the staff of Ndlovu Medical Centre and Ndlovu Research Centre for assisting in acquiring follow-up clinical and molecular diagnostic data for the patient. We would also like to thank the patient for his participation.
Author contributions. CNU and HAT conceived of and designed the study. CNU, AM and MM performed SARS-CoV-2 and respiratory panel RT-PCR, rapid antigen testing, FBC analysis, SARS-CoV-2 genotyping and SARS-CoV-2 cDNA synthesis, primer pool preparation and PCR multiplexing for SARS-CoV-2 Artic sequencing, and CNU performed data analysis. All authors contributed towards the writing of the manuscript, initially drafted by CNU, ZNM and HAT. All authors contributed towards editing and proofreading and approved the final submitted version.
Funding. This work was funded by Ndlovu Research Consortium.
Conflicts of interest. None.
References
1. Phillips SP, Wei X, Kwong JC, et al. Duration of SARS-CoV-2 shedding: A population-based, Canadian study. PLoS ONE 2021;16(6):e0252217. https://doi.org/10.1371/journal.pone.0252217 [ Links ]
2. Fontana LM, Villamagna AH, Sikka MK, McGregor JC. Understanding viral shedding of severe acute respiratory coronavirus virus 2 (SARS-CoV-2): Review of current literature. Infect Control Hosp Epidemiol 2021;42(6):659-668. https://doi.org/10.1017/ice.2020.1273 [ Links ]
3. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020;395(10229):1054-1062. https://doi.org/10.1016/S0140-6736(20)30566-3 [ Links ]
4. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and metaanalysis. Lancet Microbe 2021;2(1):e13-e22. https://doi.org/10.1016/S2666-5247(20)30172-5 [ Links ]
5. Li T-Z, Cao Z-H, Chen Y, et al. Duration of SARS-CoV-2 RNA shedding and factors associated with prolonged viral shedding in patients with COVID-19. J Med Virol 2021;93(1):506-512. https://doi.org/10.1002/jmv.26280 [ Links ]
6. Hao S, Lian J, Lu Y, et al. Decreased B cells on admission associated with prolonged viral RNA shedding from the respiratory tract in coronavirus disease 2019: A case-control study. J Infect Dis 2020;222(3):367-371. https://doi.org/10.1093/infdis/jiaa311 [ Links ]
7. Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect 2020;81(1):147-178. https://doi.org/10.1016/j.jinf.2020.03.013 [ Links ]
8. Yan D, Zhang X, Chen C, et al. Characteristics of viral shedding time in SARS-CoV-2 infections: A systematic review and meta-analysis. Front Public Health 2021;9:652842. https://doi.org/10.3389/fpubh.2021.652842 [ Links ]
9. Gombar S, Chang M, Hogan CA, et al. Persistent detection of SARS-CoV-2 RNA in patients and healthcare workers with COVID-19. J Clin Virol 2020;129:104477. https://doi.org/10.1016/jjcv.2020.104477 [ Links ]
10. Liu W-D, Chang S-Y, Wang J-T, et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect 2020;81(2):318-356. https://doi.org/10.1016/jjinf.2020.03.063 [ Links ]
11. Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol 2020;92(10):1807-1809. https://doi.org/10.1002/jmv.25941 [ Links ]
12. Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020;323(15):1502-1503. https://doi.org/10.1001/jama.2020.2783 [ Links ]
13. Kandetu T-B, Dziuban EJ, Sikuvi K, et al. Persistence of positive RT-PCR results for over 70 days in two travelers with COVID-19. Disaster Med Public Health Prep 2020;1-2. https://doi.org/10.1017/dmp.2020.450 [ Links ]
14. Widders A, Broom A, Broom J. SARS-CoV-2: The viral shedding vs infectivity dilemma. Infect Dis Health 2020;25(3):210-215. https://dolorg/10.1016/j.idh.2020.05.002 [ Links ]
15. Munker D, Osterman A, Stubbe H, et al. Dynamics of SARS-CoV-2 shedding in the respiratory tract depends on the severity of disease in COVID-19 patients. Eur Respir J 2021;58(1):2002724. https://doi.org/10.1183/13993003.02724-2020 [ Links ]
16. Mondi A, Lorenzini P, Castilletti C, et al. Risk and predictive factors of prolonged viral RNA shedding in upper respiratory specimens in a large cohort of COVID-19 patients admitted to an Italian reference hospital. Int J Infect Dis 2021;105:532-539. https://doi.org/10.1016/j.ijid.2021.02.117 [ Links ]
17. Zheng S, Fan J, Yu F, et al Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January - March 2020: Retrospective cohort study. BMJ 2020;369:m1443. https://doi.org/10.1136/bmj.m1443 [ Links ]
18. Turner JS, Day A, Alsoussi WB, et al. SARS-CoV-2 viral RNA shedding for more than 87 days in an individual with an impaired CD8+ T cell response. Front Immunol 2021;11:618402. https://doi.org/10.3389/fimmu.2020.618402 [ Links ]
19. Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021;592(7853):277-282. https://doi.org/10.1038/s41586-021-03291-y [ Links ]
20. Karim F, Moosa MYS, Gosnell BI, et al. Persistent SARS-CoV-2 infection and intra-host evolution in association with advanced HIV infection. medRxiv 2021 (epub 4 June 2021). https://doi.org/10.1101/2021.06.03.21258228 [ Links ]
21. Hartman WR, Hess AS, Connor JP. Persistent viral RNA shedding after COVID-19 symptom resolution in older convalescent plasma donors. Transfusion (Paris) 2020;60(10):2189-2191. https://doi.org/10.1111/trf.15927 [ Links ]
22. Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with coronavirus disease 2019 (COVID-19). Clin Infect Dis 2020;71(15):799-806. https://doi.org/10.1093/cid/ciaa351 [ Links ]
23. Shi D, Wu W, Wang Q, et al. Clinical characteristics and factors associated with long-term viral excretion in patients with severe acute respiratory syndrome coronavirus 2 infection: A single-center 28-day study. J Infect Dis 2020;222(6):910-918. https://doi.org/10.1093/infdis/jiaa388 [ Links ]
24. Tarhini H, Recoing A, Bridier-Nahmias A, et al. Long-term severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectiousness among three immunocompromised patients: From prolonged viral shedding to SARS-CoV-2 superinfection. J Infect Dis 2021;223(9):1522-1527. https://doi.org/10.1093/infdis/jiab075 [ Links ]
25. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020;183(7):1901-1912. https://doi.org/10.1016/j.cell.2020.10.049 [ Links ]
26. Decker A, Welzel M, Laubner K, et al Prolonged SARS-CoV-2 shedding and mild course of COVID-19 in a patient after recent heart transplantation. Am J Transplant 2020;20( 11):3239-3245. https://doi.org/10.1111/ajt.16133 [ Links ]
27. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020;383(23)2291-2293. https://doi.org/10.1056/NEJMc2031364 [ Links ]
28. Baang JH, Smith C, Mirabelli C, et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021;223(1):23-27. https://doi.org/10.1093/infdis/jiaa666 [ Links ]
29. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158(6):1831-1833. https://doi.org/10.1053/j.gastro.2020.02.055 [ Links ]
30. Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci 2021;118(21):e2105968118. https://doi.org/10.1073/pnas.2105968118 [ Links ]
31. Van Kampen JJA, van de Vijver DAMC, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021;12(1):267. https://doi.org/10.1038/s41467-020-20568-4 [ Links ]
32. Owusu D, Pomeroy MA, Lewis NM, et al. Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: A cohort study of individuals with COVID-19. J Infect Dis 2021;224(8):1362-1371. https://doi.org/10.1093/infdis/jiab107 [ Links ]
33. Zapor M. Persistent detection and infectious potential of SARS-CoV-2 virus in clinical specimens from COVID-19 patients. Viruses 2020;12(12):E1384. https://doi.org/10.3390/v12121384 [ Links ]
34. Tom MR, Mina MJ. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin Infect Dis 2020;71(16):2252-2254. https://doi.org/10.1093/cid/ciaa619 [ Links ]
 Correspondence:
Correspondence:
C N Umunnakwe
cumunnakwe@ndlovu.com
Accepted 18 October 2021














