Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
SAMJ: South African Medical Journal
versão On-line ISSN 2078-5135
versão impressa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.111 no.3 Pretoria Mar. 2021
http://dx.doi.org/10.7196/samj.2021.v111i3.15154
IN PRACTICE
ISSUES IN PUBLIC HEALTH
Qualitative and quantitative detection of SARS-CoV-2 RNA in untreated wastewater in Western Cape Province, South Africa
R JohnsonI, II; C J F MullerIII, IV, V; S GhoorVI, VII; J LouwVIII, IX; E ArcherX; S Surujlal-NaickerXI; N BerkowitzXII, XIII; M VolschenkXIV; L H L BrockerXV; G WolfaardtXVI, XVII; M van der WaltXVIII; A M MutshembeleXVIII; S MalemaXIX; H C GelderblomXX; M MdhluliXXI; G GrayXXII; A MatheeXXIII, XXIV; R A StreetXXV
IPhD; Biomedical Research and Innovation Platform, South African Medical Research Council, Cape Town, South Africa
IIPhD; Division of Medical Physiology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
IIIPhD; Biomedical Research and Innovation Platform, South African Medical Research Council, Cape Town, South Africa
IVPhD; Division of Medical Physiology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
VPhD; Department of Biochemistry and Microbiology, University of Zululand, KwaDlangezwa, South Africa
VIMTech; Biomedical Research and Innovation Platform, South African Medical Research Council, Cape Town, South Africa
VIIMTech; Genomics Centre, South African Medical Research Council, Cape Town, South Africa
VIIIPhD; Biomedical Research and Innovation Platform, South African Medical Research Council, Cape Town, South Africa
IXPhD; Department of Biochemistry and Microbiology, University of Zululand, KwaDlangezwa, South Africa
XPhD; Department of Microbiology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
XIPhD; Scientific Services, Water and Sanitation Department, City of Cape Town Metropolitan Municipality, South Africa
XIIMB ChB, MPH; City Health, City of Cape Town, South Africa
XIIIMB ChB, MPH; Department of Public Health, School of Public Health and Family Medicine, Faculty of Health Sciences, University of Cape Town, South Africa
XIVMSc; Scientific Services, Water and Sanitation Department, City of Cape Town Metropolitan Municipality, South Africa
XVMSc; Department of Microbiology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
XVIPhD; Department of Microbiology, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa
XVIIPhD; Department of Chemistry and Biology, Ryerson University, Toronto, Canada
XVIIIPhD; Tuberculosis Platform, South African Medical Research Council, Cape Town, South Africa
XIXPhD; Environment and Health Research Unit, South African Medical Research Council, Johannesburg, South Africa
XXMD, PhD, MPH; COVID-19 Prevention Network, Fred Hutchinson Cancer Research Center, Seattle, Wash., USA
XXIPhD, MBA; Chief Research Operations Office, South African Medical Research Council, Cape Town, South Africa
XXIIMB BCh, FC Paed (SA), DSc (Honoris Causa); Office of the President, South African Medical Research Council, Cape Town, South Africa
XXIIIPhD; Environment and Health Research Unit, South African Medical Research Council, Johannesburg, South Africa
XXIVPhD; Environmental Health Department, Faculty of Health Sciences, University of Johannesburg, South Africa
XXVPhD, MPH; Environment and Health Research Unit, South African Medical Research Council, Johannesburg, South Africa
ABSTRACT
Recent studies have shown that the detection of SARS-CoV-2 genetic material in wastewater may provide the basis for a surveillance system to track the environmental dissemination of this virus in communities. An effective wastewater-based epidemiology (WBE) system may prove critical in South Africa (SA), where health systems infrastructure, testing capacity, personal protective equipment and human resource capacity are constrained. In this proof-of-concept study, we investigated the potential of SARS-CoV-2 RNA surveillance in untreated wastewater as the basis for a system to monitor COVID-19 prevalence in the population, an early warning system for increased transmission, and a monitoring system to assess the effectiveness of interventions. The laboratory confirmed the presence (qualitative analysis) and determined the RNA copy number of SARS-CoV-2 viral RNA by reverse transcription polymerase chain reaction (quantitative) analysis from 24-hour composite samples collected on 18 June 2020 from five wastewater treatment plants in Western Cape Province, SA. The study has shown that a WBE system for monitoring the status and trends of COVID-19 mass infection in SA is viable, and its development and implementation may facilitate the rapid identification of hotspots for evidence-informed interventions.
Wastewater-based epidemiology (WBE) has been used effectively to monitor the circulation of viral pathogens in specific populations. WBE was originally defined as a non-intrusive tool to measure biomarkers of effect at community level. Although this approach has been extensively utilised for estimating substance use and abuse, it has recently been extended to include public health biomarkers.[1] To date, WBE has played a key role in the development of early warning systems (EWSs) for a range of enteric viruses including poliovirus, norovirus and hepatitis E.[2-4] Scientific evidence recently emerged that SARS-CoV-2, the virus that causes the disease COVID-19, is shed in faeces of infected symptomatic and asymptomatic individuals,[5,6] with detectable market RNA, ending up in wastewater.[7,8] The detection of SARS-CoV-2 RNA in faeces has stimulated rapid research into wastewater surveillance. An increasing number of countries,
including The Netherlands, Spain, Australia, the UK and the USA, have detected SARS-CoV-2 RNA in untreated wastewater.[7-12] Based on these studies and the success of previous wastewater surveillance strategies, the tracking of SARS-CoV-2 in wastewater has been proposed as a potentially important public health strategy to combat the current COVID-19 pandemic.
The first case of COVID-19 in South Africa (SA) was reported on 5 March 2020 and was followed by a national lockdown 3 weeks later. By early July 2020, over 1.5 million COVID-19 tests had been conducted, with over 130 000 confirmed cases disproportionally spread across the country[13] Currently, COVID-19 hotspots in SA are determined by human testing data which, owing to numerous factors including undetected asymptomatic cases and operational issues, may not accurately represent case numbers.[14] In SA, and other socioeconomically vulnerable countries, under-reporting of CO VID-19 cases may pose a challenge to the management of COVID-19, and an effective, complementary EWS that can be used to identify COVID-19 hotspots and emerging outbreaks will therefore be a valuable tool to guide action and the distribution of resources for containment and mitigation strategies. As viral shedding may occur before COVID-19 cases are reported,[10] WBE by molecular detection of viral RNA in wastewater can be used as a potential tool for public health monitoring at a community level. As such, this study aimed to evaluate an RNA extraction and quantitative reverse transcription polymerase chain reaction (RT-qPCR) as a means to qualitatively and quantitatively detect the presence or absence of SARS-CoV-2 viral RNA from 24-hour composite wastewater samples from wastewater treatment plants (WWTPs) in Western Cape Province, SA.
Wastewater sampling
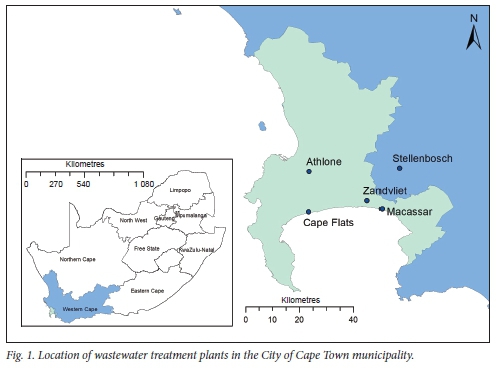
Untreated influent wastewater samples were collected on 18 June 2020 from four WWTPs in the City of Cape Town metrópole and one in the neighbouring municipality of Stellenbosch, using 24-hour time-proportional composite samplers (100 mL every 10 minutes, 09h00 - 09h00). The samples were collected at the raw influent of each WWTP after coarse screens and de-gritting (prior to discharge to primary settlement reactors and/or addition of chemical flocculants). The WWTPs (and the populations they serve) were Athlone (population -385 000), Macassar (-200 000), Cape Flats (-855 000), Zandvliet (-755 000) and Stellenbosch (-180 000) (Fig. 1). At the time of sampling, the Western Cape was the epicentre of the SARS-CoV-2 outbreak in SA. All samples were transported on ice to the laboratory and stored at -80°C before subsequent viral RNA extraction, within 24 hours, using a modified method described by Peccia et al.[10]
Viral RNA extraction from wastewater
SARS-CoV-2 viral RNA was extracted from the wastewater samples using the RNeasy PowerSoil Kit as per the manufacturer's instructions (Qiagen, Germany).[10] Briefly, 50 - 100 mL influent wastewater was spun down at 3 500 g for 20 minutes, and 5-10 mL of the pellet was added to a 15 mL PowerBead Tube containing lysis buffer, provided with the kit, to stabilise viral RNA. Thereafter, the sample was homogenised and phase-separated using an equal volume of phenol/chloroform, and the upper aqueous phase was transferred to a new 15 mL tube and mixed with buffers supplied with the RNeasy PowerSoil Kit. The aqueous phase was then transferred to the RNeasy JetStar Mini Column to elute out the bound RNA before centrifugation at 13 000 g for 10 minutes. The resultant pellet was dried and dissolved in ribonuclease-free water to a final volume of 70 µL. Total RNA was measured by spectrophotometry using a NanoDrop Spectrophotometer (ThermoFisher Scientific, USA). It should be noted that attempts to extract RNA from the supernatant yielded no detectable RNA.
Amplification of SARS-CoV-2 viral RNA using RT-qPCR analysis
Primer and probe sets, approved by the Centers for Disease Control and Prevention, that target two sequences from the SARS-CoV-2 nucleocapsid gene (N gene; Nl and N2 primer/probe) were purchased from Whitehead Scientific (2019-nCov CDC EUA Kit, Integrated DNA Technologies, USA). The N-gene primer/probe was used to assess the presence of SARS-CoV-2 viral titres. Both the Nl and N2 primer/probe sets aligned 100% to the N protein of the SARS-CoV-2 strain. The sequences of the primers and probes targeted to the N gene, including the thermocycling conditions used to amplify these regions for detection, are shown in Table 1.
Viral load quantification was assessed using the standard curve method. For RT-qPCR positive control and viral RNA copy number quantification, a 10-fold serial dilution was made using the 2019-nCoV-N-positive plasmid control as a standard, which was supplied at 200 000 copies/µL (Qauntabio, USA).[15] For the qualitative detection of SARS-CoV-2 viral RNA, a one-step RT-qPCR reaction was performed using iTaq Universal Probes One-Step Reaction Mix (Bio-Rad Laboratories, USA) in a final reaction volume of 10 uL using 1 uL of 0.1 µg/µL of total RNA. The RT-qPCR reactions were conducted on the Applied Biosystems QuantStudio 7 Flex Real-Time PCR System (ABI Technologies, SA) using universal cycling conditions as per the manufacturers instructions (Table 1). All reactions were done in duplicate and a reagent blank was included for each sample. To minimise potential contamination, RNA extraction and RT-qPCR were performed in separate laboratories.
Qualitative detection of SARS-CoV-2 viral RNA extraction using RT-qPCR analysis
During the peak of the COVID-19 outbreak in the Western Cape, total RNA ranging between 500 and 1 000 ng/mL was extracted from either 50 or 100 mL of influent wastewater using the Qiagen RNeasy PowerSoil Kit. Quantitative PCR (qPCR) analysis revealed that all samples were amplified above the quantification cycle threshold (Ct) and ranged between 29 and 32.8 cycles (samples diluted 1:10) for either the Nl or N2 primer/probe assays. A viral load of <35 cycles can be considered as a wastewater sample with low SARS-CoV-2 RNA. The 2019-nCoV_N-positive control was 10-fold serially diluted and used to determine copy numbers between the unknown wastewater sample and the calibrator. The slopes for the Nl and N2 assays were -3.4 and -3.5, respectively, and y-intercept values were -37.47 (for the Nl assay) and -37.81 (for the N2 assay). The correlation (r2) values for the Nl and N2 assays were both 0.99, while the amplification efficiency for the two assays was 94% and 95%, respectively.
Quantitative and qualitative detection of SARS-CoV-2
Quantitative and qualitative analysis using either Nl or N2 primer/ probes confirmed the presence of SARS-CoV-2 viral RNA in all five wastewater samples, with a viral RNA copy number ranging between 4.6 X 103 and 454 x 103 virual RNA copies/mL (Fig. 2A). The lowest concentration of viral RNA copy numbers was from the Stellenbosch WWTR with a Ct value of 32, while the highest was detected in the Zandvliet WWTR with a Ct value of 29 (Fig. 1). Based on the Western Cape government COVID-19 dashboard,[16] the number of confirmed COVID-19 cases on 18 June 2020 in the Khayelitsha subdistrict (one of the areas served by the Zandvliet WWTP) was higher (n=80) than in the Stellenbosch municipality (n=43). Our Zandvliet and Stellenbosch WWTP SARS-CoV-2 data corresponded with this difference in higher and lower case numbers, respectively. The COVID-19 dashboard showed that 1 week before the wastewater collection date there were 82 positive cases in Khayelitsha and 17 in Stellenbosch.

Wastewater-based epidemiology-based calculations
The calculated viral RNA copy numbers (viral copy numbers/mL) for the N1 and N2 fragments from the various WWTPs were normalised to account for the variable treatment size of the WWTPs used in this study and the variable populations that are connected to the sewer works. The average daily flow rate (in mL/day) at the raw inlet of the WWTPs was used to calculate an estimated daily load of viral RNA copy numbers for each location (in viral copy numbers/day; Fig. 2B). The population estimates from each WWTP were then used to generate an estimated per capita daily load (copy numbers/ day/1 000 inhabitants) of the viral RNA markers at the respective WWTPs (Fig. 2C). In this way, the RT-qPCR quantitative results for SARS-CoV-2 RNA markers were normalised to compensate for the variation in WWTP flow conditions received by each plant, as well as to compensate for the variable population size in each community that is served by the respective WWTPs for more accurate comparisons between locations.
What does this mean for a COVID-19 EWS?
In this proof-of-concept study, we set out to establish whether SARS-CoV-2 viral RNA could be detected both quantitatively and qualitatively in 24-hour composite wastewater samples as the basis for an EWS for COVID-19 infection in communities in urban settings of SA. The study confirmed that the Qiagen RNeasy PowerSoil Kit is a robust extraction method for the detection of SARS-CoV-2 viral RNA in wastewater. For quantitative analysis, the 2019-nCoV EUA N-gene assays produced positive results for all samples tested. The CDC-approved primer/probe was specifically designed for the detection of SARS-CoV-2 in wastewater.[15,17] Our study confirmed previous findings showing that the N gene can be used to detect SARS-Cov-2 viral RNA in wastewater samples in the USA,[11] The Netherlands,[8] the UK,[10] Spain,[9] Italy[12] and Australia.[7] Furthermore, to the best of our knowledge, this is the first published study on the viral load copy numbers from wastewater obtained from WWTPs in SA, and the first report to quantitatively detect SARS-CoV-2 viral RNA in wastewater samples from five WWTPs using the CDC Nl and N2 assays. The study further demonstrated that the viral load in the wastewater samples from Stellenbosch was much lower than that in the samples from Zandvliet (Fig. 2B and C, respectively), and this finding corresponded with the number of positive COVID-19 cases detected in the areas at the time. This is important, as tracking of SARS-CoV-2 viral RNA in wastewater could complement surveillance efforts in the development of an EWS for local COVID-19 outbreaks. As these wastewater samples were collected at the peak of the epicentre in the Western Cape, longer-term wastewater surveillance will be required to establish a baseline to distinguish between high-risk and low-risk events. Furthermore, while normalisation of data provided some insight, data on normalisation factors may be lacking in resource-constrained settings. Having baseline temporal data from which trends can be identified is therefore crucial. In this case, comparing standardised volumes (copies/mL) between sites is in fact an adequate endpoint.
An effective COVID-19 EWS may prove critical in SA and other low- and middle-income countries with inadequate health systems infrastructure, resources and human capacity. Furthermore, it can be argued that benefits of investment in a WBE system in SA will reach beyond the current COVID-19 wave, as it may forewarn of future resurgences. To date, the screening of wastewater is recognised as an important tool for monitoring wild polio-virus and vaccine-derived polioviruses,[18,19] and there are opportunities to learn from polio wastewater surveillance to overcome potential logistic and operational issues for WBE of SARS-CoV-2. As such, our future planned studies will include regular sampling coupled with SARS-CoV-2 prevalence estimation. Currently, WBE is best suited to a converging sewer network serving a target population. However, in SA, 40% of households are not connected to the sewage system.[20] Although wastewater surveillance for SARS-CoV-2 has the potential to be a powerful public health tool, especially in resource-constrained settings, alternatives to sewage from wastewater treatment plants therefore need to be supported by resolutions for more locally relevant situations (such as wastewater runoff in informal settlements) considering sanitation diversity[21] We acknowledge that this study did not include a spike RNA control to investigate the percent viral recovery, and this will be included in a follow-up study.
Conclusions
This proof-of-concept study confirms that qualitative and quantitative detection of SARS-CoV-2 viral RNA from wastewater influent samples can be used as a surveillance tool in the current SA context, thereby establishing the potential for an EWS for monitoring status and trends of COVID-19 mass infections in SA that could facilitate the rapid identification of hotspots for evidence-informed interventions. An important yet unexpected outcome was that this study brought together professionals and experts spanning skill sets, disciplines and sectors including public health, microbiology, town planning and wastewater treatment facilities, and formed the basis for a comprehensive weekly follow-up study that includes a larger number of wastewater treatment plants being sampled over consecutive days with the overall agenda of advancing equity and health.
Declaration. None.
Acknowledgements. The South African Medical Research Council is thanked for funding and support, and the City of Cape Town and Stellenbosch for provision of wastewater.
Author contributions. RJ, RAS, AM, CJFM, GG, MM conceptualised; designed, wrote and edited the manuscript. RJ and SG conducted and analysed the experiments. LHLB collected the samples. CJFM, JL, EA, SS-N, NB, MV, LHLB, GW, MvdW, GG, HCG, LHLB, SM, AMM and MM provided input towards the manuscript.
Funding South African Medical Research Council
Conflicts of interest. None.
References
1. Choi PM, Tscharke B, Samanipour S, et aL Social, demographic, and economic correlates of food and chemical consumption measured by wastewater-based epidemiology. Proc Natl Acad Sci USA 2019;116(43):21864-21873. https://doi.org/10.1073/pnas.1910242116 [ Links ]
2. Hovi T, Shulman L, van der Avoort H, Deshpande J, Roivainen M, de Gourvilie E. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol Infect 2012;140(1):1-13. https://doi.org/10.1017/S095026881000316X [ Links ]
3. Smith DB, Paddy JO, Simmonds P. The use of human sewage screening for community surveillance of hepatitis E virus in the UK. J Med Virol 2016;88(5):915-918. https://doi.org/10.1002/jmv.24403 [ Links ]
4. Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples last for 42 days. J Med Viro J 2020;92(10):1807-1809. https://doi.org/10.1002/jmv.25941 [ Links ]
5. D'Amico F, Baumgart DC, Danese S, Peyrin-Biroulet L. Diarrhea during Covid-19 infection! Pathogenesis, epidemiology, prevention and management. Clin Gastroenterol Hepatol 2020;18(8):1663-1672. https://doi.org/10.1016/j.cgh.2020.04.001 [ Links ]
6. Tiwari S, Dhole TN. Assessment of enteroviruses from sewage water and clinical samples during eradication phase of polio in North India. Virol J 2018;15(1):157-164. https://doi.org/10.1186/S12985-018-1075-7 [ Links ]
7. Ahmed W, Angel N, Edson J, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia. A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 2020;728:138764. https://doi.org/10.1016/j.scitotenv.2020.138764 [ Links ]
8. Medema G, Heijnen L, Eisinga G, ItaJiaander R, Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett 2020;7(7):511-516. https://doi.org/10.1021/acs.estlett.0c00357 [ Links ]
9. Orive G, Lertxundi U, Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci Total Environ 2020;732:139298. https://doi.org/10.1016/j.scitotenv.2020.13929? [ Links ]
10. Peccia J, Zulli A, Brackney DE, et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv 2020 (epub 12 June 2020). https://doi.org/10.1101/2020.05.19.20105999 [ Links ]
11. Sherchan SP, Shahin S, Ward LM, et al. First detection of SARS-CoV-2 RNA in wastewater in North America. A study in Louisiana, USA. Sci Total Environ 2020;743:140621. https://doi.org/10.1016/j.scitotenv.2020.140621 [ Links ]
12. La Rosa G, Iaconelli M, Mancini P, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ 2020;736:139652. https://doi.org/10.1016/j.scitotenv.2020.139652 [ Links ]
13. National Department of Health, South Africa. COVID-19 Corona Virus South African Resource Portal. 2020. https://sacoronavirus.co.za/ (accessed 17 September 2020). [ Links ]
14. Adepoju P. Nigeria responds to COVID-19. First case detected in sub-Saharan Africa. Nat Med 2020;26(4):444-448. https://doi.org/10.1038/d41591-020-00004-2 [ Links ]
15. Centers for Disease Control and Prevention. CDC 2019-novel Coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. Revision 06. 1 December 2020. https://www.fda.gov/media/134922/download (accessed 17 September 2020). [ Links ]
16. Western Cape Government. Covid-19 response. https://coronavirus.westerncape.gov.za/covid-19-dashboard (accessed 28 November 2020). [ Links ]
17. US Centers for Disease Control and Prevention. 2019-novel Coronavirus (2019-nCoV) realtime rRT-PCR panel primers and probes. Last updated 29 May 2020. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf (accessed 8 August 2020). [ Links ]
18. Gumede N, Okeibunor J, Diop O, et al. Progress on the implementation of environmental surveillance in the African Region, 2011 - 2016. J Immunol Sci 2018;S004:24-30. https://doi.org/10.29245/2578-3009/2018/SÍ.1103 [ Links ]
19. World Health Organization. Guidelines for environmental surveillance of poliovirus circulation. March 2003. https://apps.who.int/iris/bits1ream/handle/10665/67854/WHO_V-B_03.03_engpdf?sequence=l&isAUowed=y (accessed 17 September 2020). [ Links ]
20. Statistics South Africa. General Household Survey 2017. Pretoria. Stats SA, 2018. https://www.statssa.gov.za/publications/P0318/P03182017.pdf (accessed 8 August 2020). [ Links ]
21. Street R, Malema S, Mahlangeni N, Mathee A. Wastewater surveillance for Covid-19. An African perspective. Sci Total Environ 2020;743:140719. https://doi.org/10.1016%2Fj.scitotenv.2020.140715 [ Links ]
 Correspondence:
Correspondence:
R Street
renee.street@mrc.ac.za
Accepted 30 November 2020














