Serviços Personalizados
Artigo
Indicadores
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em Google
Similares em Google
Compartilhar
SAMJ: South African Medical Journal
versão On-line ISSN 2078-5135
versão impressa ISSN 0256-9574
SAMJ, S. Afr. med. j. vol.110 no.1 Pretoria Jan. 2020
http://dx.doi.org/10.7196/samj.2020.v110i1.13841
RESEARCH
Analysis and comparison of cumulative antibiograms for the Charlotte Maxeke Johannesburg Academic Hospital adult intensive care and high-care units, 2013 and 2017
T LawI, II; V ChibabhaiIII, IV; T NanaV, VI
IMB ChB, DTM&H, FC Path (SA) (Microbiol), MMed (Microbiol); Department of Clinical Microbiology and Infectious Diseases, School of Pathology, University of the Witwatersrand, Johannesburg, South Africa
IIMB ChB, DTM&H, FC Path (SA) (Microbiol), MMed (Microbiol); Clinical Microbiology Laboratory, Charlotte Maxeke Johannesburg Academic Hospital and National Health Laboratory Service, Johannesburg., South Africa
IIIMB BCh, DCH (SA), Dip HIV Man (SA), FC Path (SA) (Microbiol), MMed (Microbiol); Department of Clinical Microbiology and Infectious Diseases, School of Pathology, University of the Witwatersrand, Johannesburg, South Africa
IVMB BCh, DCH (SA), Dip HIV Man (SA), FC Path (SA) (Microbiol), MMed (Microbiol); Clinical Microbiology Laboratory, Charlotte Maxeke Johannesburg Academic Hospital and National Health Laboratory Service, Johannesburg., South Africa
VMB BCh, DTM&H, FC Path (SA) (Microbiol), MMed (Microbiol); Department of Clinical Microbiology and Infectious Diseases, School of Pathology, University of the Witwatersrand, Johannesburg, South Africa
VIMB BCh, DTM&H, FC Path (SA) (Microbiol), MMed (Microbiol); Clinical Microbiology Laboratory, Charlotte Maxeke Johannesburg Academic Hospital and National Health Laboratory Service, Johannesburg., South Africa
ABSTRACT
BACKGROUND. Infection is a common complication for patients in intensive care units (ICUs), and increasing antimicrobial resistance (AMR) is a major concern. It is therefore crucial to monitor AMR patterns in order to support clinical decision-making and antimicrobial stewardship strategies. Clinical microbiologists should provide annual cumulative antibiogram reports, which can be used to guide initial empirical antimicrobial therapy for the management of infections.
OBJECTIVES. To analyse the cumulative antibiograms for the Charlotte Maxeke Johannesburg Academic Hospital (CMJAH) combined adult multidisciplinary ICU and high-care unit (HCU) for 2013 and 2017, compare the antimicrobial susceptibility testing (AST) patterns between the 2 years, and analyse the subset of blood culture isolates.
METHODS. A retrospective descriptive analysis was performed of routine bacterial and fungal culture and AST data extracted from the National Health Laboratory Service laboratory information system for the ICU/HCU. Only the first diagnostic isolate of a given species per patient per year was included in the analysis. All analysis and reporting were done in accordance with the applicable Clinical and Laboratory Standards Institute guidelines.
RESULTS. Enterobacteriaceae predominated in first-isolate cultures in 2013 (60%) and 2017 (56%). There was an overall decrease in extended-spectrum beta-lactamase-producing Enterobacteriaceae from 2013 (42%) to 2017 (30%) (p=0.013), accompanied by an increase in carbapenem-resistant Enterobacteriaceae from 2013 (4%) to 2017 (11%) (p=0.24). Although the total percentage of Acinetobacter spp. decreased in 2017 (p=0.021), the proportion of extensively drug-resistant isolates doubled to 68% in 2017 (p<0.001). The percentage of methicillin-resistant Staphylococcus aureus decreased significantly from 49% to 14% (p<0.001), along with a significant decrease in vancomycin-resistant enterococci from 17% to 0% (p=0.001). Candida auris increased from 0% in 2013 to 11% in 2017 (p=0.002), and non-albicans Candida spp. predominated (80%) in blood cultures in 2017 (p=0.023).
CONCLUSIONS. Appropriate selection of empirical antimicrobial therapy should be guided by the ICU-specific antibiogram. The recommended empirical antimicrobial therapy at the CMJAH ICU/HCU based on the antibiogram analysis would include ertapenem to cover the Enterobacteriaceae. Amikacin is recommended for empirical treatment of suspected pseudomonal infections. Additional empirical antimicrobial therapy for Gram-positive organisms is not routinely advocated, and empirical antifungal therapy with amphotericin Β or micafungin is only appropriate in patients at high risk for invasive candidiasis.
Infection is a common complication in patients in intensive care units (ICUs) and is associated with considerable mortality and morbidity, and increased costs.[1] Antimicrobial treatment of patients with sepsis is increasingly complicated by the alarming rates of antimicrobial resistance (AMR) among pathogens. The ICU is often called the epicentre of AMR development owing to its extremely vulnerable population, with increased risks of becoming infected through multiple procedures and use of invasive devices.[2-4] Most large epidemiological studies of infection and sepsis in ICUs have been conducted in Europe, North America and Australia, with limited data from southern Africa.[1] With increasing AMR worldwide, it is crucial to monitor emerging trends in AMR at the local level to support clinical decision-making, infection control interventions and antimicrobial stewardship (AMS) strategies.[5,6] The most urgent and serious threats for the ICU include infections with extended-spectrum beta-lactamase-producing (ESBL) Enterobacteriaceae, derepressed AmpC beta-lactamases and carbapenemases, extensively drug-resistant (XDR) and carbapenem-resistant Acinetobacter baumannii, multidrug-resistant (MDR) Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus (MRSA), and azole-resistant and MDR Candida spp.[7,8]
In its guide for the prevention of hospital-acquired infections, the World Health Organization[9] specifies that clinical microbiologists are responsible for providing annual reports of antimicrobial susceptibility patterns of pathogens. Epidemiological surveillance activities by microbiology laboratories are therefore growing in importance.[10,11] Monitoring of AMR trends is commonly performed in healthcare facilities using an annual summary of susceptibility rates, known as a cumulative antibiogram.[5,10] The most frequent use of a cumulative antibiogram report is to guide initial empirical antimicrobial therapy for the management of infections in patients who do not yet have definitive microbiological results to target treatment. The choice of empirical antimicrobial coverage is critical in the ICU because initiation of inadequate empirical therapy has been associated with poor clinical outcomes.[3,12] Bloodstream infections represent a common complication among critically ill patients in the ICU, and a leading cause of morbidity and mortality Early appropriate antibiotic therapy is therefore a critically important aspect of the treatment of these patients.[13] Cumulative antibiograms have additional applications, including updating periprocedural or perioperative prophylaxis recommendations, providing a rationale for antimicrobial formulary selection, surveying local resistance patterns, and identifying targets for AMS and best practices.[10,14]
No cumulative antibiogram studies have yet been published from the Charlotte Maxeke Johannesburg Academic Hospital (CMJAH) adult ICU and high-care unit (HCU), or any other ICUs in South Africa (SA). This study was designed to provide the necessary cumulative antibiograms for the CMJAH ICU/HCU, and to demonstrate AMR changes and trends between two time periods. In 2017 the SA National Department of Health mandated the implementation of AMS interventions at healthcare facilities in order to combat the emerging threat of AMR. Emergence of antibiotic resistance is highly correlated with selective pressure resulting from excessive use of antimicrobials in the ICU. It is therefore crucial for ICU clinicians to have regularly updated antibiograms in order to make informed decisions about empirical antibiotic choices.
Objectives
The primary study objectives were to prepare and analyse the cumulative antibiograms for the ICU/HCU for the years 2013 and 2017, and to compare the different organisms isolated and their antimicrobial susceptibility testing (AST) patterns between the 2 years. The secondary objective was to specifically analyse the cumulative antibiogram data for the subset of blood culture isolates.
Methods
Study setting
CMJAH is an academic tertiary-level hospital with a 12-bed multidisciplinary ICU and an 8-bed HCU. Adult patients admitted to these units frequently have severe sepsis and multiple organ dysfunction with or without septic shock, or have recently undergone complex surgery.[15]
Study design
A retrospective descriptive analysis of all the routine bacterial and fungal culture and AST data from the CMJAH ICU/HCU was performed. All the data were extracted from the National Health Laboratory Service (NHLS) laboratory information system (LIS). Culture and AST data from 1 January 2013 to 31 December 2013 were compared with data from 1 January 2017 to 31 December 2017. Clinical samples were tested at the NHLS microbiology laboratory based at CMJAH. The laboratory used a variety of identification and AST methodologies, which included the manual Kirby-Bauer disc diffusion test and the Etest (bioMérieux, France), as well as the automated Microscan (Beckman Coulter Inc., USA) and Vitek 2 (bioMérieux, France). AST results were interpreted according to the contemporary Clinical and Laboratory Standards Institute (CLSI) guidelines.[16,17] Molecular confirmation of carbapenemase production was performed at the Antimicrobial Resistance Laboratory at the National Institute for Communicable Diseases from 2014 onwards.
CLSI guideline CLSI M39-A4[14] was used to guide the compilation of the cumulative antibiograms, as it provides criteria for standardising and benchmarking antibiograms. To eliminate the bias inherent in an 'all-isolates' approach, only the first diagnostic isolate of a given species per patient per analysis period was included, as this approach has direct relevance to guiding recommendations for initial empirical therapy. Culture and susceptibility reports from samples collected for surveillance or screening were excluded.
Definitions
First isolate refers to the initial microbial isolate of a particular species recovered from a patient during the time period analysed, regardless of body source, specimen type or AST profile.[14]
Susceptible refers to a category where isolates are inhibited by the usually achievable concentrations of antimicrobial agent when the dose recommended to treat the site of infection is used.[14]
Susceptible dose dependent means that the isolate's minimum inhibitory concentration is high, but increased dosing of the agent has the potential to inhibit the yeast in vivo.[14]
Non-susceptible is a category used for isolates that are not inhibited by the usually achievable concentrations of antimicrobial agent with the normal dosage schedules, and includes intermediate and resistant.[14]
AmpC. For the Enterobacteriaceae, non-susceptibility to cefoxitin was used as a marker of inducible ampicillin class C beta-lactamase production.[18]
ESBL. Non-susceptibility to third- and/or fourth-generation cephalosporins was used to predict ESBL production.'[16,17]
CRE. Non-susceptibility to any of the carbapenems was a marker of carbapenem-resistant Enterobacteriaceae (CRE). Non-susceptibility to carbapenems may be the result of various mechanisms, including production of carbapenemases or combinations of AmpC, ESBL and porin loss.[16,17]
CPE. Only Enterobacteriaceae with a confirmed carbapenemase-producing gene were defined as carbapenemase-producing Enterobacteriaceae (CPE).[16,17]
MDR and XDR. Definitions of MDR and XDR were applied from Magiorakos et al. [19] to report on the AST resistance profiles of Acinetobacter spp. and Pseudomonas spp.
Statistical analysis
Statistical analyses were performed using Excel version 2010 (Microsoft Corporation, USA) andGraphPad version 8.0 (Graphpad Software, USA). Categorical data were presented as percent susceptibility for each antimicrobial agent tested. The Agresti-Coull method was used to calculate confidence intervals, and Fisher's exact test to compare differences between the two observed percent susceptible estimates from 2013 v. 2017. The p-values were reported as two-tailed, and values <0.05 were considered statistically significant.
Ethical approval
The study was approved by the Human Research Ethics Committee, University of the Witwatersrand (ref. no. W-CBP-180802-3).
Results
Analysis of first isolates
Of the 594 first-isolate cultures in 2013, 53% (n=314) were Gram-negative bacteria, 33% (n=196) were Gram-positive bacteria and 14% (n=84) were Candida spp. In 2017, 59% (n=388) of the 662 first-isolate cultures were Gram-negative bacteria, 30% (n=200) were Gram-positive bacteria and 11% (n=74) were Candida spp. The increase in the proportion of Gram-negative bacteria from 53% in 2013 to 59% in 2017 was statistically significant (p=0.046). Numbers of anaerobic organisms isolated in both 2013 (Bacteroides spp., n=2) and 2017 (Bacteroides spp., n=12) were low. Table 1 shows the distribution of all the first isolates by culture site.
Enterobacteriaceae
The Enterobacteriaceae made up the largest proportion of first-isolate cultures in 2013 (60%) and 2017 (56%) (Table 2). Among the Enterobacteriaceae isolated in 2013 and 2017, there were three main genera: Klebsiella spp., Escherichia spp. and Enterobacter spp. In the panel of antimicrobial agents tested, the only significant change was an overall decrease in susceptibility to piperacillin-tazobactam from 75% in 2013 to 64% in 2017 (p=0.017). The statistically significant decrease in ESBL-producing isolates from 42% in 2013 to 30% in 2017 (p=0.013) was accompanied by a significant increase in the proportion of CREfrom 4% in 2013 to 11% in 2017 (p=0.024). Only 4 of the 8 CRE were sent for genotyping in 2013, of which 1 was a blaVIM, 1 was a blaIMP, and 2 tested negative for carbapenemase genes. In 2017, 21 of the 23 CRE were sent for genotyping. The predominant carbapenamase was bla0XA_4Sand its variants (n=17). Three blaNDMand 1 ΜαΏΜwere also detected.
Klebsiella spp. were the most frequent Gram-negative bacteria isolated in both years. In 2013, 61% of all Klebsiella spp. were ESBL producers, and this decreased to 34% in 2017 (p=0.001) owing to the accompanying increase in CRE from 6% in 2013 to 18% in 2017 (p=0.029). In comparison with 2013, Klebsiella spp. demonstrated higher susceptibility to amoxicillin-clavulanic acid (p=0.008), cefepime (p=0.03) and trimethoprim-sulfamethoxazole (p=0.006) in 2017 (Fig. 1A). Of the aminoglycosides, Klebsiella spp. were most susceptible to amikacin in both years. In 2013 and 2017, >50% of the Klebsiella spp. were non-susceptible to third-generation cephalosporins. Klebsiella spp. were all susceptible to tigecycline in 2017, but no comparison could be made with 2013 as tigecycline was not tested.
Of the Enterobacteriaceae, E. coli was the second most prevalent organism isolated in both years (Table 2). Susceptibility to the third- and fourth-generation cephalosporins was >60% (Fig. 1B). Amikacin was the most susceptible aminoglycoside for E. coli. The only antimicrobial agents with susceptibilities <50% in both years were ampicillin, amoxicillin-clavulanic acid and trimethoprim-sulfamethoxazole.
Enterobacter spp. were the third most frequently isolated Enterobacteriaceae in the study years. Enterobacter spp. produce an inducible AmpC beta-lactamase, and expression of this enzyme confers resistance to broad-spectrum cephalosporins including cefotaxime, ceftazidime and ceftriaxone. More than 50% of the isolates were susceptible to the fourth-generation cephalosporin cefepime (Fig. 1C). Again, amikacin was the most susceptible aminoglycoside for the Enterobacter spp.
The remaining Enterobacteriaceae that were isolated had <30 isolates per genus, and were therefore grouped together as other Enterobacteriaceae' for the purposes of analysis. In this group, >80% of isolates were susceptible to the aminoglycosides, as well as ciprofloxacin, in both 2013 and 2017 (Fig. 1D).
Non-fermentative Gram-negative bacteria
In 2013, Acinetobacter spp. were the predominant non-fermenters (52%), followed by Pseudomonas spp. (42%) (Table 2). In contrast, in 2017, Pseudomonas spp. predominated (44%), followed by Acinetobacter spp. (38%). The remaining non-fermentative Gram-negative bacteria isolated had numbers <30 per genus. However, owing to their intrinsic MDR, they were included in the analysis. In 2013, 5 Stenotrophamonas maltophilia and 2 Burkholderia cepacia were isolated, and in 2017, 21 Burkholderia spp. and 6 S. maltophilia were isolated.
The overall reduction in the number of Acinetobacter spp. in 2017 was statistically significant (p=0.021). In 2013, 62% were MDR compared with only 14% in 2017 (p<0.0001). This decrease was due to the significant doubling of the percentage of XDR Acinetobacter spp., from 34% in 2013 to 68% in 2017 (p<0.001). There was a significant increase in non-susceptibility in 2017 to ceftazidime (p=0.008), gentamicin (p=0.016) and tobramycin (p<0.001) (Fig. 2A). In 2017, >60% of all antimicrobials tested were non-susceptible for Acinetobacter spp., and susceptibility to the carbapenems, meropenem and imipenem, was <20%. Tigecycline was susceptible in 89% of Acinetobacter spp. in 2017, but no comparison could be made with 2013 as tigecycline was not tested.
There were limited changes in Pseudomonas spp. between the 2 study years. A significant reduction in the susceptibility of piperacillin-tazobactam was observed, from 92% in 2013 to 78% in 2017 (p=0.04) (Fig. 2B). There was a significant increase in the number of Burkholderia spp. from 2013 to 2017 (p<0.001). Both S. maltophilia and Burkholderia spp. had very high non-susceptibility rates to multiple antimicrobials (>69%) (Fig. 2C), which is in keeping with their intrinsically resistant phenotype.
Gram-positive bacteria
Of the 196 Gram-positive bacteria isolated in 2013, 58% were Staphylococcus spp. and 37% were Enterococcus spp. (Table 3). Similarly, in 2017, of the 200 Gram-positive bacterial isolates, 64% were Staphylococcus spp. and 28% were Enterococcus spp. For the remaining 5% (n=10) of Gram-positive organisms isolated in 2013 and 8% (n=16) in 2017, AST analysis was omitted owing to the small number of isolates.
Staphylococcus spp. made up the largest proportion of the first-isolate Gram-positive cultures in both 2013 (n=114) and 2017 (n=128). The proportion of MRSA decreased significantly in 2017 to 14% (p<0.001). Additionally, the susceptibility of S. aureus to rifampicin also increased significantly in 2017 (p=0.005) (Fig. 3A). All S. aureus isolates were susceptible to vancomycin and linezolid in both study years. The vast majority of methicillin-resistant coagulase-negative staphylococci (CoNS) were resistant to cloxacillin. However, there was a significant reduction in the proportion of methicillin-resistant CoNS, from 83% in 2013 to 72% in 2017 (p=0.016) (Fig. 3B). The susceptibility of CoNS to trimethoprim-sulfamethoxazole also increased significantly in 2017 (p=0.028).
E. faecium predominated in both years, and consequently only one-third of Enterococcus spp. were susceptible to ampicillin (Fig. 3C).
In 2013, 17% of the Enterococcus spp. isolated were vancomycin-resistant enterococci (VRE). This reduced significantly in 2017, when no VRE were isolated (p=0.001). All the Enterococcus spp. in both study years were susceptible to linezolid.
Candida spp.
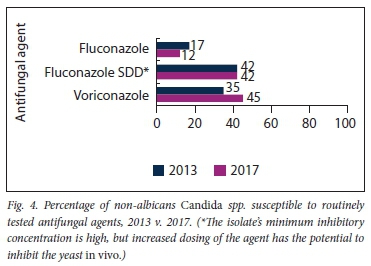
Table 4 summarises the different Candida spp. isolated in both the study years. C. albicans was the predominant species isolated in both 2013 (64%) and 2017 (65%). C. albicans remained 100% susceptible to the two routinely tested azole antifungals, fluconazole and voriconazole, in both years. There were two significant changes in the species distribution in the study years: C. parapsilosis decreased from 7% in 2013 to 0% in 2017 (p=0.030), and C. auris increased from 0% in 2013 to 11% in 2017 (p=0.002). Overall susceptibility of non-albicans Candida spp. (NAC) to the azole antifungals remained <50% for both years (Fig. 4). Micafungin and amphotericin Β were 100% susceptible in 2017, but no comparison could be made with 2013, when these antifungals were not tested.
Analysis of bloodstream isolates
Table 5 summarises the first-isolate organisms from blood cultures. Of the Gram-negative blood culture isolates, the Enterobacteriaceae predominated in both 2013 (62%) and 2017 (55%). All the members of the Enterobacteriaceae isolated each year were analysed together as a group to allow for sufficient numbers. Klebsiella spp., E. coli and Enterobacter spp. were found to be the predominant species in both years. There was an overall decrease in ESBL-producing Enterobacteriaceae from 52% in 2013 to 32% in 2017 (p=0.032). Of all the antimicrobial agents tested, there was only a significant increase in susceptibility for gentamicin, from 41% in 2013 to 63% in 2017 (p=0.013).
All the non-fermentative Gram-negative bacteria isolated each year were analysed together as a group to allow for sufficient numbers. In 2013, 29% of the non-fermenters were MDR compared with 9% in 2017 (p=0.005); however, there was a non-significant increase in the percentage of XDR non-fermenters from 29% in 2013 to 37% in 2017. A significant decrease in the susceptibility of tobramycin from 54% in 2013 to 19% in 2017 (p=0.005) was noted, and there was a significant increase in the proportion of Burkholderia spp. from 3% in 2013 to 24% in 2017 (p=0.009).
Of the Gram-positive bacteria isolated from blood cultures, CoNS predominated in both 2013 (54%) and 2017 (60%). There was a significant reduction in the proportion of methicillin-resistant CoNS, from 83% in 2013 to 62% in 2017 (p=0.040).
Of all the Candida spp. isolated from blood cultures in 2013, C. albicans was the predominant species isolated (58%); however, in 2017 the NAC predominated (80%), and this difference was statistically significant (p=0.023). In keeping with this change, there was a significant reduction in overall susceptibility of the bloodstream Candida spp. to fluconazole, from 63% in 2013 to 20% in 2017 (p=0.01).
Discussion
This is the first study describing the cumulative antibiogram results for the CMJAH multidisciplinary adult ICU and HCU. The findings from this study provide important epidemiological information. The pertinent findings include the predominance of Enterobacteriaceae in 2013 and 2017. Of concern was the overall increase in CRE, XDR A. baumannii and Burkholderia spp. The significant reduction in both MRSA and VRE is noteworthy. Among the Candida spp. isolated, the emergence of MDR C. auris and a predominance of NAC in bloodstream isolates in 2017 reflects current global epidemiology[20]
The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus,Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) represent the six most common MDR pathogens threatening patients in ICUs globally.[21] These pathogens were also prevalent bacterial isolates in the CMJAH ICU/HCU. The predominance of Enterobacteriaceae in this unit is similar to the findings of the global EPIC II study.[1] The decrease in the proportion of ESBL-producing Enterobacteriaceae was coupled with a significant increase in the proportion of CRE from 4% to 11%, a finding that could not be corroborated by other studies. The majority of the CPE (65%) were Klebsiella spp., and the predominant carbapenamase was bla and its variants (81%), which is in keeping with published national surveillance data from SA.[22,23] The number of blaOXA48 isolates may underestimate the true prevalence in this unit, as blaOXA48 isolates could have appeared susceptible to carbapenems on routine AST and would therefore not have been sent for genotyping.[22]
Also in keeping with published data, Klebsiella spp. were the most frequent Gram-negative bacteria isolated in both years.[23] Despite the overall ESBL decrease, Klebsiella spp. remained the predominant ESBL-producing Enterobacteriaceae in both years. The reasons for the increase in susceptibility of three antimicrobials in 2017, amoxicillin-clavulanic acid, cefepime, and trimethoprim-sulfamethoxazole, are not known. The CLSI cefepime breakpoints were lowered between 2013 and 2017, so an increase in cefepime resistance would have been expected over the study years.[16,17] Strain typing of the Klebsiella spp. between the different time points may be a useful tool to assist with understanding changes in antibiograms, but this would require isolates to be stored for prospective analysis. Imipenem and meropenem had the highest susceptibilities (97-98%) of all the antimicrobial agents tested in both years. Ertapenem still had high susceptibility rates (93%) and would therefore be the most appropriate choice for empirical treatment of Enterobacteriaceae in cases of suspected nosocomial infection. For patients in septic shock with a suspected CRE infection, combination empirical therapy is associated with improved survival.[24] The addition of amikacin would be recommended, as among the CRE in 2017 specifically, 83% (19/23) of the isolates were susceptible to amikacin.
The overall prevalence of non-fermentative Gram-negative bacteria did not differ between the study years. There was, however, a significant increase in Burkholderia spp. over the 2 years, with most isolates (71%) being recovered from blood culture. This trend has also been seen elsewhere in ICU patients without cystic fibrosis.[25] There was a surprising decrease in the overall percentage of Acinetobacter spp. These bacteria can be found in the natural environment as well as occurring as commensals of the skin and body secretions. The true prevalence of infection caused by Acinetobacter spp. is difficult to assess, as there are no guidelines to assist in differentiating between isolates that cause infection v. colonisation.[26] The increasing resistance of Acinetobacter spp. from 2013 to 2017 was evidenced by the doubling of XDR isolates in 2017. This increasing resistance is also in keeping with local and international published data.[23,26]
Susceptibility patterns of Pseudomonas spp. remained stable over the 2 study years, with a significant decrease only in the susceptibility of piperacillin-tazobactam, to <80%, making this agent inappropriate for the empirical therapy of nosocomial Pseudomonas spp. infections. The carbapenems were the least susceptible antimicrobials tested for Pseudomonas spp. Amikacin had the highest susceptibility (94%) of all the antimicrobial agents tested in 2017, and would therefore be the agent of choice for empirical treatment of suspected nosocomial pseudomonal infections.
Susceptibility of Acinetobacter spp. to the carbapenems was very low (<20%), but remained constant, and susceptibility was highest to tigecycline in 2017. For the non-fermenters on blood cultures, there was a very high non-susceptibility rate to the panel of antimicrobials tested, and in 2017, the most susceptible agent was ceftazidime at 62%. Empirical therapy with meropenem or imipenem will not provide adequate cover for either Acinetobacter spp. or Pseudomonas spp. For Acinetobacter spp. infections assessed as clinically significant optimal therapy will probably require the use of Colistin for patients with septic shock. However, reliable Colistin AST was not available during the study years.[23] New AST methods for Colistin (broth microdilution) have been implemented in the laboratory, and Colistin AST results would need to be studied going forward.
Among the Gram-positive isolates, Staphylococcus spp. predominated in both study years. CoNS are normal skin commensals and are frequently isolated from clinical specimens. Determining whether an isolate of CoNS represents a true infection or colonisation is very difficult, and there are no simple criteria with sufficient specificity to assist with this decision.[27] On a positive note, the proportion of MRSA and VRE isolates decreased significantly. A similar decline in MRSA was also reported in another SA surveillance study[23,28] CoNS are commonly implicated in catheter-related bloodstream infections, and the CMJAH ICU/HCU treats these infections by removal of the infected catheter, without the use of vancomycin. Empirical use of vancomycin is therefore infrequent in this unit, which may have played a role in the decreased rates of MRSA and VRE. In 2017, all the Gram-positive cocci isolated were 99 - 100% susceptible to vancomycin and linezolid, making these agents appropriate for empirical therapy when Gram-positive organisms are suspected to be significant.
Although C. albicans remained the predominant yeast isolated overall, in both 2013 (64%) and 2017 (65%) it could have been considered a commensal in many cultures (e.g. the respiratory tract urine and skin). There was a concerning increase in the percentage of MDR C. amis in 2017, from 0% to 17%. This is consistent with the dramatic increase of C. amis elsewhere in SA and globally over the past 4 years.[20]Candida spp. made up the minority of blood culture isolates in both years. However, as evidenced by numerous studies, blood cultures have low sensitivity, despite being the gold standard for the definitive diagnosis of candidaemia.[29] C. albicans was the predominant species isolated (58%) from blood cultures in 2013, but of concern was that NAC predominated (80%) in 2017, which is in keeping with the global epidemiological shift of Candida spp.[20] As a result of this shift in epidemiology, the azoles are no longer the agents of choice for empirical therapy of Candida spp. infections. Susceptibilities to micafungin and amphotericin Β were 100% in 2017. These antifungals should be considered the agents of choice for empirical antifungal therapy in patients at high risk of invasive candidiasis, depending on the local availability and guideline recommendation.
This study provides the necessary/required cumulative antibiogram data for the CMJAH ICU/HCU, which can be used by clinicians to guide their empirical selection of antimicrobial therapy. The current NHLS LIS was not primarily designed as a research or surveillance tool,[30] so computer software limitations and data extraction methods need to be refined further in order to make the provision of annual cumulative antibiograms an achievable task by the clinical microbiology laboratory.
Study limitations
There were several limitations to this study. Firstly, because of the retrospective nature of the data analysis, real-time changes in AST or emergence of resistance are not reflected. Secondly, clinical data were not available to distinguish between hospital-acquired and community-acquired infections, and true infection v. colonisation. Thirdly, in order to reduce the bias that may be present in an all-isolates approach, a first-isolate approach was used, as recommended by CLSI. This approach may, however, underestimate the resistance rate of nosocomial infections. Fourthly, the laboratory changed automated AST methods from Microscan in 2013 to Vitek 2 in 2017, and this could have affected AST results. In addition, the study was not able to analyse or report on Colistin AST, as new testing methods were recommended in 2017 by CLSI and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), and these had not yet been routinely implemented by the microbiology laboratory during the study years. Lastly, tigecycline, amphotericin Β and micafungin were not tested in 2013, so no comparison could be made for these antimicrobials.
Conclusions
Management of infections in ICU patients is an evolving challenge because of the ever-present threat of resistant isolates. The appropriate selection of empirical antibiotic therapy should be guided by ICU-specific antibiograms. Based on this unit's antibiogram, empirical antimicrobial therapy should always cover the Enterobacteriaceae, and the agent of choice would be ertapenem. Amikacin is recommended for empirical treatment of suspected pseudomonal infections. Additional empirical antimicrobial therapy for the Gram-positive organisms is not routinely advocated, as the majority of isolates in this study were CoNS. In 2017, most S. aureus isolated were methicillin susceptible. Ertapenem is active against methicillin-sensitive S. aureus and will offer coverage in the setting of empirical use. There was a low incidence of culture-proven candidaemia in this study, so empirical antifungal therapy with amphotericin Β or micafungin would only be appropriate in patients at high risk of invasive candidiasis with accompanying clinical signs suggestive of such infections.
In order to adequately implement AMS as a tool to combat AMR in ICUs nationally, further prospective multicentre epidemiological studies are needed at multidisciplinary ICUs across SA.
Declaration. This publication is a result of the work done by TL for her MMed (Microbiol) degree.
Acknowledgements. None.
Author contributions. The research question was conceived by TL. TL performed the data collection and was the primary author. VC and TN contributed editing and supervision of the writing of the article.
Funding. This study was supported by the Department of Clinical Microbiology and Infectious Diseases, University of the Witwatersrand.
Conflicts of interest. None.
References
1. Vincent J-L, Rello J, Marshall J, etat International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009;302(21).2323-2329. https://doi.org/10.1001/jama.2009.1754 [ Links ]
2. Vincent J-L, Bassetti M, Francois B, et al Advances in antibiotic therapy in the critically ill. Crit Care 2016;20(1)133. https://doi.org/10.1186/s13054-016-1285-6 [ Links ]
3. Campion M, Scully G. Antibiotic use in the intensive care unit. Optimization and de-escalation. J Intensive Care Med 2018;33(12)647-655. https://doi.org/10.1177/0885066618762747 [ Links ]
4. Brusselaers N, Vogelaers D, Blot S. The rising problem of antimicrobial resistance in the intensive care unit. Ann Intensive Care 2011;1.47. https://doi.org/10.1186/2110-5820-1-47 [ Links ]
5. Hindier JF, Stelling J. Analysis and presentation of cumulative antibiograms. A new consensus guideline from the Clinical and Laboratory Standards Institute. Clin Infect Dis 2007;44(6).867-873. https://doi.org/10.1086/511864 [ Links ]
6. Kollef MH, Bassetti M, Francois B, et al. The intensive care medicine research agenda on multidrug-resistant bacteria, antibiotics, and stewardship. Intensive Care Med 2017;43(9).1187-1197. https://doi.org/10.1007/s00134-017-4682-7 [ Links ]
7. Bassetti M, de Waele JJ, Eggimann P, etal. Preventive and therapeutic strategies in critically ill patients with highly resistant bacteria. Intensive Care Med 2015;11(5).776-795. https://doi.org/10.1007/S00134-015-3719-Z [ Links ]
8. Chowdhary A, Sharma C, Meis JF Candida auris. A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally PLoS Pathog 2017;13(5).e1006290. https://doi.org/10.1371/journal.ppat.1006290 [ Links ]
9. World Health Organization. Prevention of Hospital-Acquired Infections. A Practical Guide. 2nd ed. Geneva, Switzerland. WHO, 2002. https://apps.who.int/iris/bitstream/handle/10665/67350/WHO_CDS_CSR_EPH_2002.12.pdf (accessed 25 November 2019). [ Links ]
10. Morency-Potvin P, Schwartz DN, Weinstein RA. Antimicrobial stewardship. How the microbioloffv laboratory can right the ship. Clin Microbiol Rev 2017;30(1).381-407. http://doi.org/10.1128/CMR.00066-16 [ Links ]
11. AmbrettiS, GagliottiC, Luzzaro F, et al Reporting epidemiology of antibiotic resistance. Microbiologia Medica 2015;30(2).34-40. https://doi.org/10.4081/mm.2015.5308 [ Links ]
12. Binkley S, Fishman NO, LaRosa LA, et al Comparison of unit-specific and hospital-wide antibiograms. Potential implications for selection of empirical antimicrobial therapy Infect Control Hosp Epidemiol 2006;27(7).682-687. https://doi.org/10.1086/505921 [ Links ]
13. Bassetti M, Right E, Carnelutti A. Bloodstream infections in the intensive care unit. Virulence 2016;7(3):267-279. https://doi.org/10.1080/21505594.2015.1134072 [ Links ]
14. Hindier JA, Barton M, Erdman SM, et al. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Testing Data. Approved Guideline. 4th ed. CLSI document M39-A4. Wayne, Perm.: Clinical and Laboratory Standards Institute, 2014. [ Links ]
15. Johnston D, Khan R Miot J, Moch S, van Deventer Y, Richards G. Usage of antibiotics in the intensive care units of an academic tertiary-level hospital. South Afr J Infect Dis 2018;33(4).106-113. https://doi.org/10.1080/23120053.2018.1482645 [ Links ]
16. Performance Standards for Antimicrobial Susceptibility Testing Twenty-Third Informational Supplement CLSI document M100-S23. Wayne, Perm.. Clinical and Laboratory Standards Institute, 2013. [ Links ]
17. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, Penn.. Clinical and Laboratory Standards Institute, 2017. [ Links ]
18. Jacoby GA. AmpC ß-lactamases. Clin Microbiol Rev 2009;22(1).161-182. https://doi.org/10.1128/CMR.00036-08 [ Links ]
19. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and ρ and rug-resistant bacteria. An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18(3).268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x [ Links ]
20. Govender NP, Magobo RE, Mpembe R, et al. Candida auris in South Africa, 2012 - 2016. Emerg Infect Dis 2018;24(11):2036-2040. https://doi.org/10.3201/eid2411.180368 [ Links ]
21. Ziiahi G, Artigas A, Martin-Loeches I. What's new in multidrug-resistant pathogens in the ICU? Ann Intensive Care 2016;6:96. https://doi.org/10.1186/s13613-016-0199-4 [ Links ]
22. Singh-Moodley A, Perovic O. Phenotypic and genotypic correlation of carbapenemase-producing Enterobacteriaceae and problems experienced in routine screening. S Afr Med J 2018;108(6).495-501. https://doi.org/10.7196/SAMJ.2018.vl08i6.12878 [ Links ]
23. Perovic O, Ismail H, van Schalkwyk E. Antimicrobial resistance surveillance in the South African public sector. South Afr J Infect Dis 2018;33(4).118-129. https://doi.org/10.1080/23120053.2018.1469851 [ Links ]
24. Gutierrez-Gutierrez B, Salamanca E, de Cueto M, et aL Effect of appropriate combination therapy on mortality of patients with bloodstream infections dueto carbapenemase-producingEnterobacteriaceae (INCREMENT). A retrospective cohort study. Lancet Infect Dis 2017;17(7).726-734. https://doi.org/10.1016/S1473-3099(17)30228-1 [ Links ]
25. Bressler AM, Kaye KS, LiPuma JJ, et ai. Risk factors for Burkholderia cepacia complex bacteremia among intensive care unit patients without cystic fibrosis. A case-control study. Infect Control Hosp Epidemiol 2007;28(8)951-958. https://doi.org/10.1086/519177 [ Links ]
26. Ogutlu A, Guclu E, Karabay O, Utku AC, Tuna N, Yahyaoglu M. Effects of carbapenem consumption on the prevalence of Acinetobacter infection in intensive care unit patients. Ann Clin Microbiol Antimicrob 2014;13.7. https://doi.org/10.1186/1476-0711-13-7 [ Links ]
27. Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am 2009;23(1).73-98. https://doi.org/10.1016/j.idc.2008.10.001 [ Links ]
28. Perovic O, Iyaioo S, Kularatne R, et ai. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010 to 2012. Laboratory based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One 2015;10(12).e0145429. https://doi.org/10.1371/journai.pone.0145429 [ Links ]
29. Clancy CJ, Nguyen M. Non-culture diagnostics for invasive candidiasis. Promise and unintended consequences. J Fungi 2018;4(1).27. https://doi.org/10.3390/jof4010027 [ Links ]
30. Nyasulu P, Paszko C, Mbelie N. A narrative review of the laboratory information system and its role in antimicrobial resistance surveillance in South Africa. Adv Microbiol 2014;4(10).692-696. https://doi.org/10.4236/aim.2014.410074 [ Links ]
 Correspondence:
Correspondence:
T Law
tina.law.za@gmail.com
Accepted 15 August 2019














